You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000604_01505
Basic Information
help
| Species |
UBA1829 sp900549415
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; UBA1829; UBA1829 sp900549415
|
| CAZyme ID |
MGYG000000604_01505
|
| CAZy Family |
PL27 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000604 |
3604218 |
MAG |
Madagascar |
Africa |
|
| Gene Location |
Start: 154801;
End: 156798
Strand: -
|
No EC number prediction in MGYG000000604_01505.
| Family |
Start |
End |
Evalue |
family coverage |
| PL27 |
50 |
650 |
5.6e-154 |
0.9491803278688524 |
MGYG000000604_01505 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 5NO8_A
|
3.66e-113 |
9 |
655 |
33 |
693 |
PolysaccharideLyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NO8_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
| 5NOK_A
|
5.31e-108 |
9 |
655 |
33 |
693 |
PolysaccharideLyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838],5NOK_B Polysaccharide Lyase BACCELL_00875 [Bacteroides cellulosilyticus DSM 14838] |
This protein is predicted as OTHER
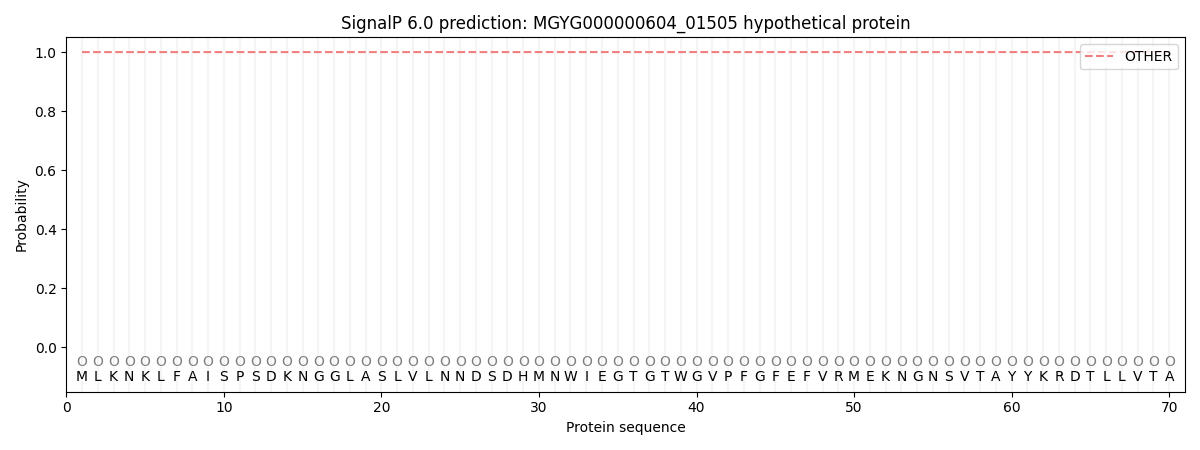
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
1.000031
|
0.000001
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
There is no transmembrane helices in MGYG000000604_01505.

