You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000605_01695
You are here: Home > Sequence: MGYG000000605_01695
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; | |||||||||||
| CAZyme ID | MGYG000000605_01695 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 130; End: 2451 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 91 | 373 | 2.3e-80 | 0.9930795847750865 |
| CBM23 | 503 | 666 | 3.3e-47 | 0.9938271604938271 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3934 | COG3934 | 1.54e-21 | 89 | 388 | 24 | 289 | Endo-1,4-beta-mannosidase [Carbohydrate transport and metabolism]. |
| pfam03425 | CBM_11 | 8.09e-16 | 504 | 666 | 7 | 173 | Carbohydrate binding domain (family 11). |
| cd14256 | Dockerin_I | 2.76e-12 | 707 | 763 | 1 | 57 | Type I dockerin repeat domain. Bacterial cohesin domains bind to a complementary protein domain named dockerin, and this interaction is required for the formation of the cellulosome, a cellulose-degrading complex. The cellulosome consists of scaffoldin, a noncatalytic scaffolding polypeptide, that comprises repeating cohesion modules and a single carbohydrate-binding module (CBM). Specific calcium-dependent interactions between cohesins and dockerins appear to be essential for cellulosome assembly. This subfamily represents type I dockerins, which are responsible for anchoring a variety of enzymatic domains to the complex. |
| pfam00150 | Cellulase | 9.33e-10 | 87 | 375 | 23 | 270 | Cellulase (glycosyl hydrolase family 5). |
| pfam03442 | CBM_X2 | 2.42e-07 | 404 | 489 | 2 | 83 | Carbohydrate binding domain X2. This domain binds to cellulose and to bacterial cell walls. It is found in glycosyl hydrolases and in scaffolding proteins of cellulosomes (multiprotein glycosyl hydrolase complexes). In the cellulosome it may aid cellulose degradation by anchoring the cellulosome to the bacterial cell wall and by binding it to its substrate. This domain has an Ig-like fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADL52789.1 | 5.30e-201 | 27 | 727 | 604 | 1361 |
| BAV13033.1 | 5.30e-201 | 27 | 727 | 604 | 1361 |
| ADZ85047.1 | 3.21e-185 | 41 | 495 | 636 | 1097 |
| QEH70547.1 | 1.01e-182 | 41 | 495 | 636 | 1097 |
| AEY66038.1 | 6.87e-144 | 37 | 406 | 30 | 395 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1RH9_A | 6.46e-51 | 43 | 381 | 6 | 344 | ChainA, endo-beta-mannanase [Solanum lycopersicum] |
| 4QP0_A | 1.06e-42 | 43 | 372 | 4 | 325 | CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
| 3PZ9_A | 4.81e-40 | 48 | 371 | 18 | 340 | Nativestructure of endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZG_A I222 crystal form of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 [Thermotoga petrophila RKU-1],3PZI_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with beta-D-glucose [Thermotoga petrophila RKU-1],3PZM_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with three glycerol molecules [Thermotoga petrophila RKU-1],3PZN_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with citrate and glycerol [Thermotoga petrophila RKU-1],3PZO_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 in complex with three maltose molecules [Thermotoga petrophila RKU-1],3PZQ_A Structure of the hyperthermostable endo-1,4-beta-D-mannanase from Thermotoga petrophila RKU-1 with maltose and glycerol [Thermotoga petrophila RKU-1] |
| 6TN6_A | 4.17e-38 | 48 | 371 | 1 | 326 | X-raystructure of the endo-beta-1,4-mannanase from Thermotoga petrophila [Thermotoga petrophila RKU-1] |
| 3WH9_A | 3.81e-32 | 43 | 389 | 2 | 324 | Theligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger],3WH9_B The ligand-free structure of ManBK from Aspergillus niger BK01 [Aspergillus niger] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FZ29 | 3.23e-54 | 39 | 377 | 25 | 367 | Mannan endo-1,4-beta-mannosidase 1 OS=Arabidopsis thaliana OX=3702 GN=MAN1 PE=2 SV=1 |
| Q6Z310 | 4.03e-54 | 25 | 372 | 20 | 367 | Putative mannan endo-1,4-beta-mannosidase 9 OS=Oryza sativa subsp. japonica OX=39947 GN=MAN9 PE=2 SV=2 |
| Q9SG94 | 9.09e-54 | 23 | 381 | 11 | 375 | Mannan endo-1,4-beta-mannosidase 3 OS=Arabidopsis thaliana OX=3702 GN=MAN3 PE=2 SV=1 |
| Q7Y223 | 9.74e-53 | 43 | 373 | 42 | 379 | Mannan endo-1,4-beta-mannosidase 2 OS=Arabidopsis thaliana OX=3702 GN=MAN2 PE=2 SV=1 |
| Q9M0H6 | 1.76e-52 | 42 | 372 | 40 | 377 | Mannan endo-1,4-beta-mannosidase 5 OS=Arabidopsis thaliana OX=3702 GN=MAN5 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
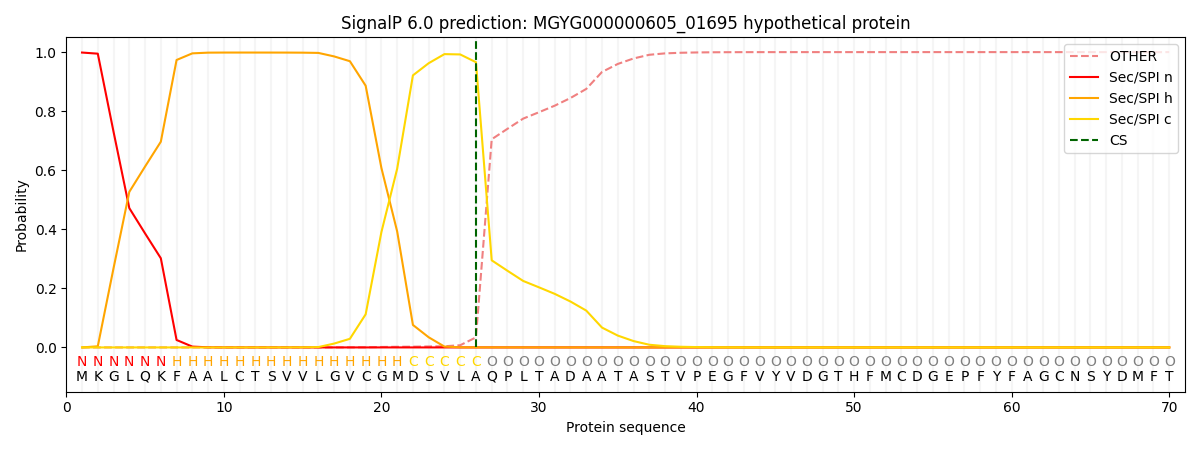
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000351 | 0.996725 | 0.002304 | 0.000220 | 0.000201 | 0.000164 |
