You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000605_01898
You are here: Home > Sequence: MGYG000000605_01898
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Ruminococcaceae; Ruminococcus_C; | |||||||||||
| CAZyme ID | MGYG000000605_01898 | |||||||||||
| CAZy Family | CBM4 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 861; End: 3032 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 286 | 712 | 3.7e-106 | 0.9880382775119617 |
| CBM4 | 34 | 159 | 5.4e-33 | 0.9920634920634921 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 3.84e-90 | 289 | 710 | 2 | 370 | Glycosyl hydrolase family 9. |
| cd02850 | E_set_Cellulase_N | 8.90e-26 | 197 | 281 | 2 | 86 | N-terminal Early set domain associated with the catalytic domain of cellulase. E or "early" set domains are associated with the catalytic domain of cellulases at the N-terminal end. Cellulases are O-glycosyl hydrolases (GHs) that hydrolyze beta 1-4 glucosidic bonds in cellulose. They are usually categorized into either exoglucanases, which sequentially release terminal sugar units from the cellulose chain, or endoglucanases, which also attack the chain internally. The N-terminal domain of cellulase may be related to the immunoglobulin and/or fibronectin type III superfamilies. These domains are associated with different types of catalytic domains at either the N-terminal or C-terminal end and may be involved in homodimeric/tetrameric/dodecameric interactions. Members of this family include members of the alpha amylase family, sialidase, galactose oxidase, cellulase, cellulose, hyaluronate lyase, chitobiase, and chitinase, among others. |
| pfam02927 | CelD_N | 7.13e-25 | 195 | 275 | 1 | 83 | Cellulase N-terminal ig-like domain. |
| PLN02613 | PLN02613 | 3.30e-13 | 284 | 708 | 25 | 469 | endoglucanase |
| PLN03009 | PLN03009 | 3.37e-13 | 278 | 710 | 21 | 481 | cellulase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16782.1 | 0.0 | 1 | 710 | 1 | 711 |
| ADZ82408.1 | 5.43e-226 | 20 | 716 | 22 | 723 |
| AQR94654.1 | 1.53e-225 | 11 | 710 | 13 | 717 |
| AGF55906.1 | 3.06e-225 | 11 | 710 | 13 | 717 |
| QEH68101.1 | 4.50e-225 | 14 | 716 | 16 | 723 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3EZ8_A | 3.66e-94 | 197 | 709 | 9 | 523 | CrystalStructure of endoglucanase Cel9A from the thermoacidophilic Alicyclobacillus acidocaldarius [Alicyclobacillus acidocaldarius subsp. acidocaldarius],3GZK_A Structure of A. Acidocaldarius Cellulase CelA [Alicyclobacillus acidocaldarius subsp. acidocaldarius],3H2W_A Structure of A. acidocaldarius cellulase CelA in complex with cellobiose [Alicyclobacillus acidocaldarius subsp. acidocaldarius],3H3K_A Structure of A. acidocaldarius cellulase CelA in complex with cellotetraose [Alicyclobacillus acidocaldarius subsp. acidocaldarius],3RX5_A structure of AaCel9A in complex with cellotriose-like isofagomine [Alicyclobacillus acidocaldarius subsp. acidocaldarius],3RX7_A Structure of AaCel9A in complex with cellotetraose-like isofagomine [Alicyclobacillus acidocaldarius subsp. acidocaldarius],3RX8_A structure of AaCel9A in complex with cellobiose-like isofagomine [Alicyclobacillus acidocaldarius subsp. acidocaldarius] |
| 5E2J_A | 8.94e-94 | 197 | 709 | 32 | 546 | Crystalstructure of single mutant thermostable endoglucanase (D468A) from Alicyclobacillus acidocaldarius [Alicyclobacillus acidocaldarius subsp. acidocaldarius],5E2J_B Crystal structure of single mutant thermostable endoglucanase (D468A) from Alicyclobacillus acidocaldarius [Alicyclobacillus acidocaldarius subsp. acidocaldarius] |
| 4CJ0_A | 2.79e-92 | 193 | 710 | 26 | 540 | ChainA, ENDOGLUCANASE D [Acetivibrio thermocellus],4CJ1_A Chain A, ENDOGLUCANASE D [Acetivibrio thermocellus] |
| 1CLC_A | 3.95e-92 | 193 | 710 | 40 | 554 | ChainA, ENDOGLUCANASE CELD; EC: 3.2.1.4 [Acetivibrio thermocellus] |
| 3X17_A | 1.54e-90 | 197 | 710 | 18 | 549 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23658 | 1.77e-94 | 197 | 710 | 4 | 538 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
| P0C2S4 | 1.53e-91 | 193 | 710 | 26 | 540 | Endoglucanase D (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celD PE=1 SV=1 |
| A3DDN1 | 2.76e-91 | 193 | 710 | 50 | 564 | Endoglucanase D OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celD PE=1 SV=1 |
| A7LXT3 | 1.80e-79 | 200 | 712 | 35 | 573 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH9A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02649 PE=1 SV=1 |
| A3DCH1 | 7.38e-79 | 13 | 709 | 7 | 803 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celK PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
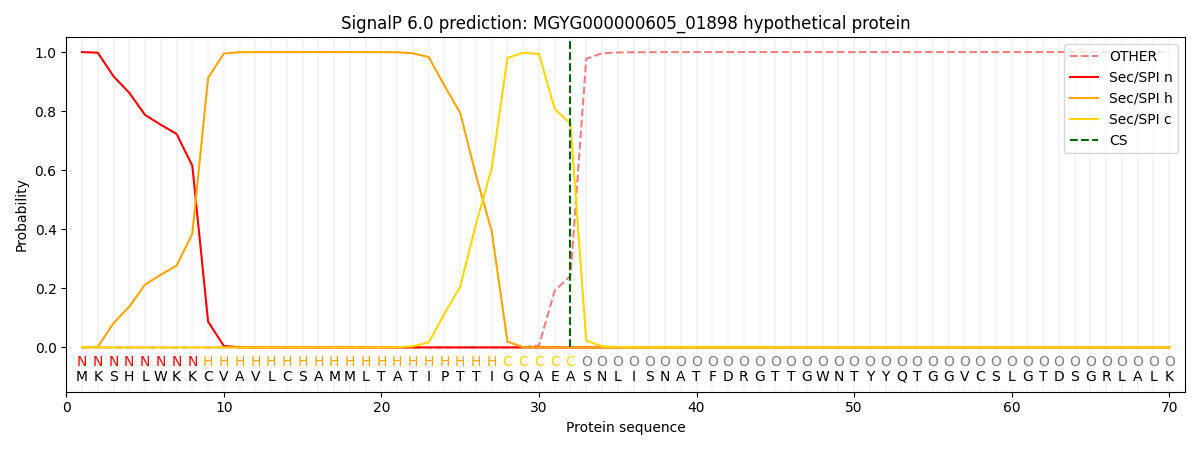
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000286 | 0.998985 | 0.000191 | 0.000199 | 0.000178 | 0.000159 |
