You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000606_00655
You are here: Home > Sequence: MGYG000000606_00655
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000433355 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000433355 | |||||||||||
| CAZyme ID | MGYG000000606_00655 | |||||||||||
| CAZy Family | GH50 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 300301; End: 303429 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH50 | 66 | 690 | 8.2e-147 | 0.9754977029096478 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam14606 | Lipase_GDSL_3 | 1.00e-53 | 859 | 1033 | 2 | 177 | GDSL-like Lipase/Acylhydrolase family. |
| cd01844 | SGNH_hydrolase_like_6 | 1.26e-40 | 860 | 1033 | 1 | 177 | SGNH_hydrolase subfamily. SGNH hydrolases are a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the Ser-His-Asp(Glu) triad found in other serine hydrolases. |
| pfam14607 | GxDLY | 2.88e-38 | 705 | 850 | 2 | 146 | N-terminus of Esterase_SGNH_hydro-type. This domain lies upstream of SGNH hydrolase, but its function is not known. There is a highly conserved GxDLY sequence-motif. |
| pfam13472 | Lipase_GDSL_2 | 7.64e-12 | 863 | 1021 | 1 | 173 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| cd00229 | SGNH_hydrolase | 3.66e-07 | 861 | 1032 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGY53973.1 | 9.83e-253 | 19 | 694 | 20 | 685 |
| AXP07840.1 | 7.53e-214 | 36 | 694 | 56 | 719 |
| ASV75458.1 | 4.62e-167 | 104 | 696 | 97 | 684 |
| QNN24957.1 | 2.08e-163 | 116 | 696 | 615 | 1213 |
| AXP07838.1 | 1.45e-159 | 115 | 696 | 141 | 712 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4BQ2_A | 9.82e-77 | 202 | 695 | 196 | 748 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ2_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_C Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ3_D Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4BQ4_A | 2.53e-76 | 202 | 695 | 196 | 748 | Structuralanalysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ4_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_A Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40],4BQ5_B Structural analysis of an exo-beta-agarase [Saccharophagus degradans 2-40] |
| 4LHS_A | 2.86e-76 | 700 | 1038 | 3 | 337 | Crystalstructure of a putative GDSL-like lipase (BACOVA_00914) from Bacteroides ovatus ATCC 8483 at 1.40 A resolution [Bacteroides ovatus ATCC 8483] |
| 6XJ9_A | 1.18e-75 | 210 | 690 | 223 | 759 | Structureof PfGH50B [Pseudoalteromonas fuliginea],6XJ9_B Structure of PfGH50B [Pseudoalteromonas fuliginea] |
| 5Z6P_A | 1.62e-75 | 116 | 690 | 129 | 759 | Thecrystal structure of an agarase, AgWH50C [Agarivorans gilvus],5Z6P_B The crystal structure of an agarase, AgWH50C [Agarivorans gilvus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P48840 | 2.78e-85 | 68 | 693 | 291 | 952 | Beta-agarase B OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaB PE=3 SV=1 |
| P48839 | 2.36e-54 | 59 | 723 | 251 | 950 | Beta-agarase A OS=Vibrio sp. (strain JT0107) OX=47913 GN=agaA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
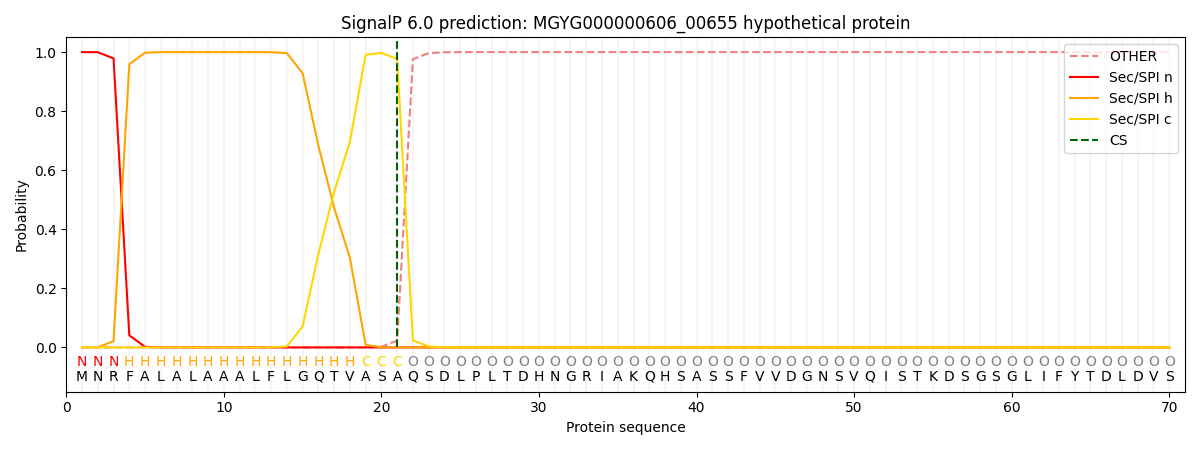
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000325 | 0.998807 | 0.000234 | 0.000204 | 0.000206 | 0.000181 |
