You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000608_00862
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; UMGS1518;
|
| CAZyme ID |
MGYG000000608_00862
|
| CAZy Family |
GH39 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 734 |
|
82827.55 |
4.6899 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000608 |
2613041 |
MAG |
Madagascar |
Africa |
|
| Gene Location |
Start: 1569;
End: 3773
Strand: +
|
No EC number prediction in MGYG000000608_00862.
| Family |
Start |
End |
Evalue |
family coverage |
| GH39 |
154 |
347 |
6.9e-18 |
0.45011600928074247 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| pfam18989
|
DUF5722 |
5.53e-04 |
228 |
292 |
118 |
176 |
Family of unknown function (DUF5722). This is a family of unknown function mainly found in bacteria. |
| COG3693
|
XynA |
7.91e-04 |
168 |
298 |
67 |
187 |
Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
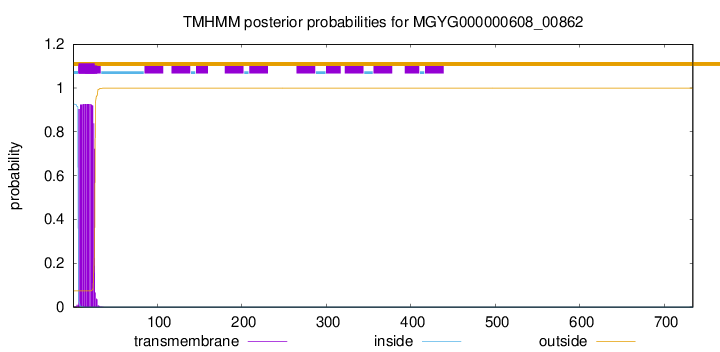
This protein is predicted as SP
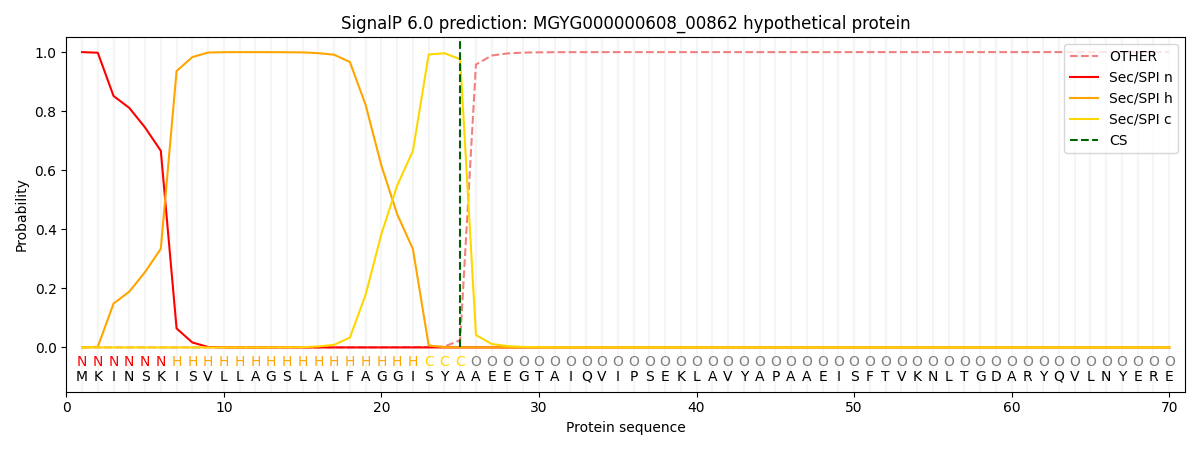
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000429
|
0.998723
|
0.000229
|
0.000220
|
0.000189
|
0.000169
|