You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000621_02121
You are here: Home > Sequence: MGYG000000621_02121
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Acetatifactor; | |||||||||||
| CAZyme ID | MGYG000000621_02121 | |||||||||||
| CAZy Family | CBM61 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 14607; End: 18713 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH53 | 466 | 846 | 2.1e-119 | 0.9970760233918129 |
| CBM61 | 938 | 1078 | 1.6e-23 | 0.9929078014184397 |
| CBM61 | 219 | 359 | 1.6e-16 | 0.9716312056737588 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3867 | GanB | 1.25e-125 | 447 | 858 | 21 | 403 | Arabinogalactan endo-1,4-beta-galactosidase [Carbohydrate transport and metabolism]. |
| pfam07745 | Glyco_hydro_53 | 1.78e-114 | 466 | 846 | 1 | 333 | Glycosyl hydrolase family 53. This domain belongs to family 53 of the glycosyl hydrolase classification. These enzymes are enzymes are endo-1,4- beta-galactanases (EC:3.2.1.89). The structure of this domain is known and has a TIM barrel fold. |
| cd00118 | LysM | 3.82e-10 | 1324 | 1367 | 3 | 45 | Lysin Motif is a small domain involved in binding peptidoglycan. LysM, a small globular domain with approximately 40 amino acids, is a widespread protein module involved in binding peptidoglycan in bacteria and chitin in eukaryotes. The domain was originally identified in enzymes that degrade bacterial cell walls, but proteins involved in many other biological functions also contain this domain. It has been reported that the LysM domain functions as a signal for specific plant-bacteria recognition in bacterial pathogenesis. Many of these enzymes are modular and are composed of catalytic units linked to one or several repeats of LysM domains. LysM domains are found in bacteria and eukaryotes. |
| smart00257 | LysM | 1.10e-08 | 1324 | 1367 | 2 | 44 | Lysin motif. |
| pfam07532 | Big_4 | 1.28e-08 | 868 | 921 | 6 | 57 | Bacterial Ig-like domain (group 4). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in a variety of bacterial surface proteins. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNM03783.1 | 0.0 | 1 | 1368 | 1 | 1357 |
| CBK73963.1 | 3.51e-229 | 251 | 1087 | 76 | 852 |
| ADL34775.1 | 4.65e-200 | 449 | 1080 | 245 | 874 |
| QGH37103.1 | 4.99e-178 | 449 | 1088 | 174 | 799 |
| ALS74893.1 | 5.65e-178 | 453 | 1085 | 43 | 658 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1R8L_A | 1.61e-140 | 444 | 852 | 3 | 394 | Thestructure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1R8L_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis [Bacillus licheniformis],1UR0_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR0_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_A The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],1UR4_B The structure of endo-beta-1,4-galactanase from Bacillus licheniformis in complex with two oligosaccharide products. [Bacillus licheniformis],2CCR_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2CCR_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_A Structure of Beta-1,4-Galactanase [Bacillus licheniformis],2J74_B Structure of Beta-1,4-Galactanase [Bacillus licheniformis] |
| 2GFT_A | 1.19e-139 | 444 | 852 | 3 | 394 | ChainA, Glycosyl Hydrolase Family 53 [Bacillus licheniformis],2GFT_B Chain B, Glycosyl Hydrolase Family 53 [Bacillus licheniformis] |
| 7OSK_A | 9.43e-78 | 455 | 849 | 40 | 394 | ChainA, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230],7OSK_B Chain B, Arabinogalactan endo-1,4-beta-galactosidase [Ignisphaera aggregans DSM 17230] |
| 1HJQ_A | 5.05e-43 | 468 | 775 | 6 | 301 | Structureof two fungal beta-1,4-galactanases: searching for the basis for temperature and pH optimum. [Humicola insolens] |
| 1FHL_A | 1.01e-35 | 468 | 775 | 6 | 302 | CrystalStructure Of Beta-1,4-galactanase From Aspergillus Aculeatus At 293k [Aspergillus aculeatus],1FOB_A Crystal Structure Of Beta-1,4-galactanase From Aspergillus Aculeatus At 100k [Aspergillus aculeatus],6Q3R_A ASPERGILLUS ACULEATUS GALACTANASE [Aspergillus aculeatus],6Q3R_B ASPERGILLUS ACULEATUS GALACTANASE [Aspergillus aculeatus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q65CX5 | 2.07e-139 | 444 | 852 | 28 | 419 | Endo-beta-1,4-galactanase OS=Bacillus licheniformis (strain ATCC 14580 / DSM 13 / JCM 2505 / CCUG 7422 / NBRC 12200 / NCIMB 9375 / NCTC 10341 / NRRL NRS-1264 / Gibson 46) OX=279010 GN=ganB PE=1 SV=1 |
| O07013 | 2.06e-134 | 455 | 852 | 43 | 423 | Endo-beta-1,4-galactanase OS=Bacillus subtilis (strain 168) OX=224308 GN=ganB PE=1 SV=1 |
| P48843 | 1.81e-49 | 465 | 776 | 6 | 314 | Uncharacterized protein in bgaB 5'region (Fragment) OS=Niallia circulans OX=1397 PE=3 SV=1 |
| P83691 | 2.77e-42 | 468 | 775 | 6 | 301 | Arabinogalactan endo-beta-1,4-galactanase OS=Humicola insolens OX=34413 PE=1 SV=1 |
| P48841 | 4.74e-39 | 440 | 775 | 3 | 330 | Arabinogalactan endo-beta-1,4-galactanase OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=ganB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
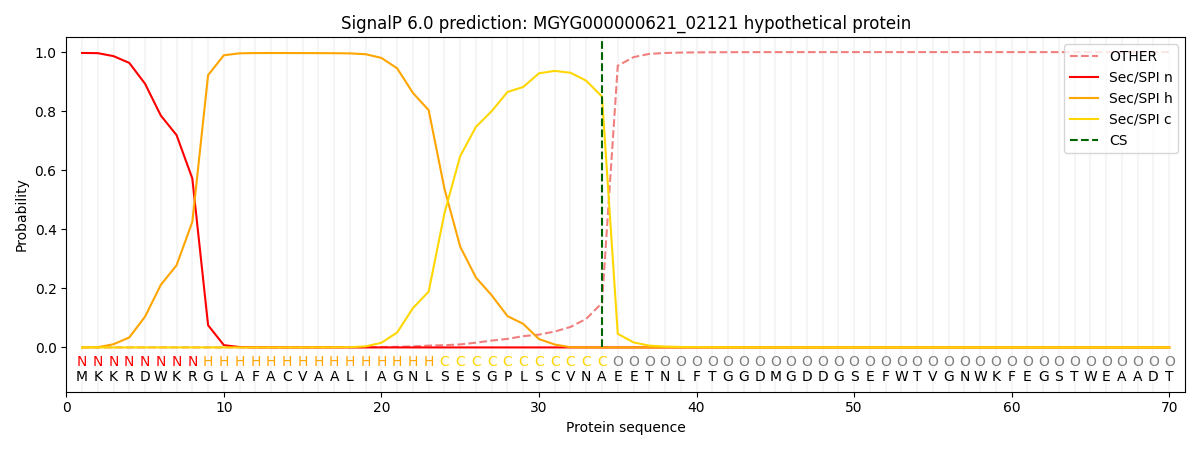
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000669 | 0.995275 | 0.003388 | 0.000272 | 0.000198 | 0.000171 |
