You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000622_01361
You are here: Home > Sequence: MGYG000000622_01361
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA636 sp900546285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; UBA636; UBA636 sp900546285 | |||||||||||
| CAZyme ID | MGYG000000622_01361 | |||||||||||
| CAZy Family | GH9 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 9040; End: 11100 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH9 | 257 | 672 | 1.7e-72 | 0.9880382775119617 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00759 | Glyco_hydro_9 | 2.30e-42 | 259 | 667 | 1 | 367 | Glycosyl hydrolase family 9. |
| PLN02308 | PLN02308 | 7.50e-11 | 302 | 609 | 70 | 411 | endoglucanase |
| PLN02420 | PLN02420 | 9.38e-11 | 299 | 611 | 80 | 433 | endoglucanase |
| PLN02613 | PLN02613 | 2.38e-10 | 298 | 667 | 64 | 468 | endoglucanase |
| PLN00119 | PLN00119 | 8.64e-10 | 301 | 611 | 72 | 415 | endoglucanase |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CBL16782.1 | 1.14e-73 | 146 | 685 | 179 | 726 |
| QEH67467.1 | 6.03e-70 | 171 | 677 | 69 | 594 |
| ADZ81800.1 | 2.93e-69 | 170 | 677 | 65 | 591 |
| AUK47910.1 | 1.14e-66 | 162 | 675 | 193 | 721 |
| ACZ98604.1 | 2.95e-66 | 65 | 682 | 59 | 727 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4CJ0_A | 1.46e-45 | 153 | 675 | 12 | 545 | ChainA, ENDOGLUCANASE D [Acetivibrio thermocellus],4CJ1_A Chain A, ENDOGLUCANASE D [Acetivibrio thermocellus] |
| 1CLC_A | 1.74e-45 | 153 | 675 | 26 | 559 | ChainA, ENDOGLUCANASE CELD; EC: 3.2.1.4 [Acetivibrio thermocellus] |
| 3X17_A | 1.00e-40 | 171 | 670 | 19 | 549 | Crystalstructure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium],3X17_B Crystal structure of metagenome-derived glycoside hydrolase family 9 endoglucanase [uncultured bacterium] |
| 5U2O_A | 1.18e-35 | 196 | 668 | 18 | 530 | Crystalstructure of Zn-binding triple mutant of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
| 5U0H_A | 3.52e-32 | 196 | 668 | 18 | 530 | Crystalstructure of GH family 9 endoglucanase J30 [Thermobacillus composti KWC4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P23658 | 3.92e-49 | 170 | 675 | 4 | 543 | Cellodextrinase OS=Butyrivibrio fibrisolvens OX=831 GN=ced1 PE=1 SV=1 |
| P0C2S4 | 8.02e-45 | 153 | 675 | 12 | 545 | Endoglucanase D (Fragment) OS=Acetivibrio thermocellus OX=1515 GN=celD PE=1 SV=1 |
| A3DDN1 | 1.07e-44 | 153 | 675 | 36 | 569 | Endoglucanase D OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celD PE=1 SV=1 |
| A7LXT3 | 5.03e-31 | 172 | 678 | 34 | 579 | Xyloglucan-specific endo-beta-1,4-glucanase BoGH9A OS=Bacteroides ovatus (strain ATCC 8483 / DSM 1896 / JCM 5824 / BCRC 10623 / CCUG 4943 / NCTC 11153) OX=411476 GN=BACOVA_02649 PE=1 SV=1 |
| P0C2S1 | 1.85e-23 | 172 | 668 | 216 | 802 | Cellulose 1,4-beta-cellobiosidase OS=Acetivibrio thermocellus OX=1515 GN=celK PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
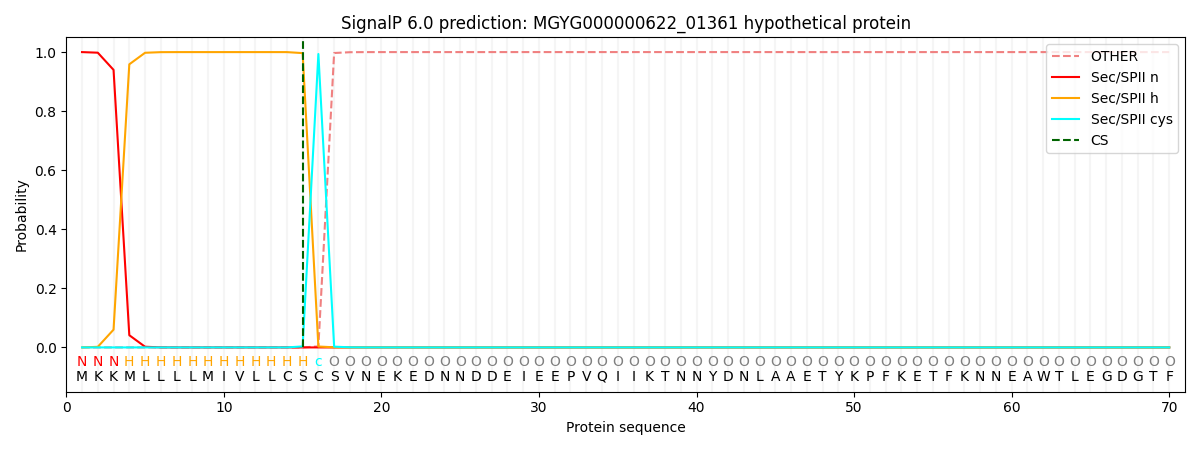
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000032 | 0.000000 | 0.000000 | 0.000000 |
