You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000626_01156
You are here: Home > Sequence: MGYG000000626_01156
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
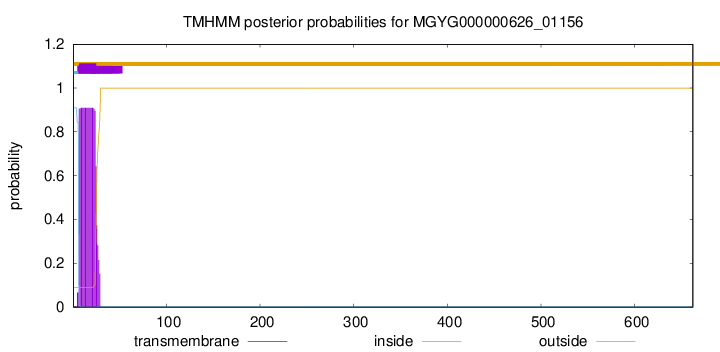
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; Eubacterium_J; | |||||||||||
| CAZyme ID | MGYG000000626_01156 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 4740; End: 6728 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 78 | 311 | 2.8e-29 | 0.9312977099236641 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 4.69e-13 | 61 | 285 | 14 | 246 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIB28168.1 | 4.60e-147 | 43 | 337 | 4 | 298 |
| QUF84801.1 | 1.43e-138 | 43 | 336 | 28 | 321 |
| QMW91197.1 | 2.85e-138 | 43 | 336 | 28 | 321 |
| BBK76626.1 | 2.85e-138 | 43 | 336 | 28 | 321 |
| AXB85103.1 | 8.06e-138 | 43 | 336 | 28 | 321 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4U5I_A | 4.35e-20 | 44 | 350 | 76 | 360 | ChainA, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U5I_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U5K_A Chain A, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U5K_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
| 4U3A_A | 1.87e-19 | 44 | 350 | 76 | 360 | ChainA, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],4U3A_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
| 3AZR_A | 5.28e-19 | 44 | 325 | 3 | 302 | DiverseSubstrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose [Thermotoga maritima MSB8],3AZR_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose [Thermotoga maritima MSB8],3AZS_A Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose [Thermotoga maritima MSB8],3AZS_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose [Thermotoga maritima MSB8],3AZT_A Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_B Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_C Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8],3AZT_D Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose [Thermotoga maritima MSB8] |
| 5BYW_A | 2.14e-17 | 44 | 350 | 76 | 371 | ChainA, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_B Chain B, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_C Chain C, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_D Chain D, Endoglucanase H [Acetivibrio thermocellus ATCC 27405],5BYW_E Chain E, Endoglucanase H [Acetivibrio thermocellus ATCC 27405] |
| 3RJY_A | 3.54e-17 | 36 | 329 | 3 | 313 | CrystalStructure of Hyperthermophilic Endo-beta-1,4-glucanase in complex with substrate [Fervidobacterium nodosum Rt17-B1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16218 | 4.60e-18 | 44 | 350 | 327 | 611 | Endoglucanase H OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=celH PE=1 SV=1 |
| P25472 | 2.55e-16 | 5 | 282 | 2 | 272 | Endoglucanase D OS=Ruminiclostridium cellulolyticum (strain ATCC 35319 / DSM 5812 / JCM 6584 / H10) OX=394503 GN=celCCD PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
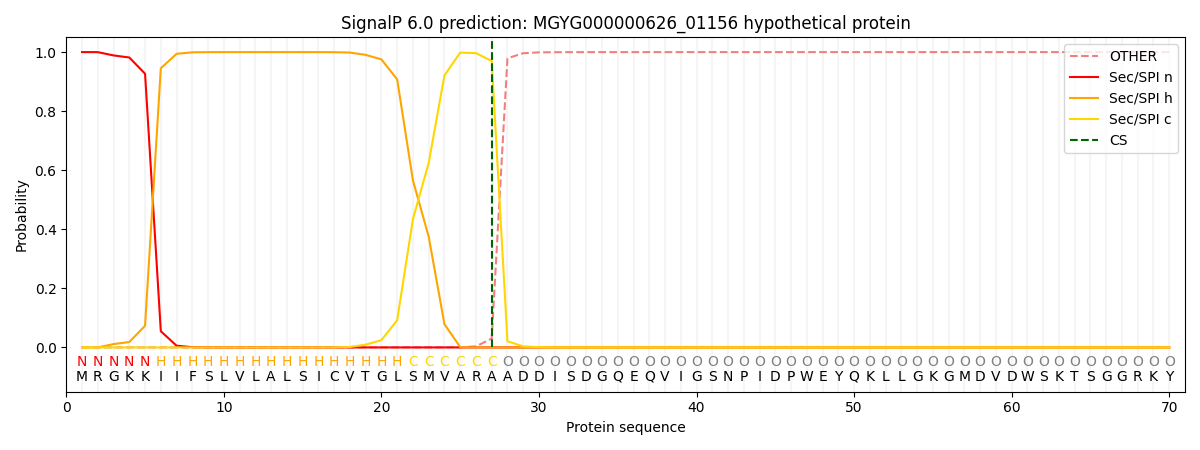
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000246 | 0.999091 | 0.000203 | 0.000170 | 0.000143 | 0.000133 |