You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000631_00193
You are here: Home > Sequence: MGYG000000631_00193
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp002299635 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp002299635 | |||||||||||
| CAZyme ID | MGYG000000631_00193 | |||||||||||
| CAZy Family | GH51 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 20494; End: 22791 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH51 | 411 | 762 | 3.5e-55 | 0.6761904761904762 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam06964 | Alpha-L-AF_C | 1.33e-18 | 612 | 759 | 1 | 192 | Alpha-L-arabinofuranosidase C-terminal domain. This family represents the C-terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase (EC:3.2.1.55). This catalyzes the hydrolysis of nonreducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| smart00813 | Alpha-L-AF_C | 6.84e-18 | 612 | 759 | 1 | 189 | Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides. |
| COG3534 | AbfA | 5.93e-12 | 414 | 522 | 33 | 172 | Alpha-L-arabinofuranosidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AGB28163.1 | 1.07e-284 | 2 | 763 | 3 | 843 |
| VEH15278.1 | 9.91e-228 | 22 | 763 | 25 | 853 |
| BCS84266.1 | 4.26e-226 | 1 | 761 | 21 | 873 |
| QNT66325.1 | 3.07e-191 | 22 | 720 | 37 | 848 |
| QVJ81530.1 | 2.53e-171 | 27 | 759 | 30 | 838 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6ZPS_AAA | 1.74e-31 | 239 | 709 | 2 | 555 | ChainAAA, MgGH51 [Meripilus giganteus],6ZPV_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPW_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPX_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPY_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZPZ_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ0_AAA Chain AAA, MgGH51 [Meripilus giganteus],6ZQ1_AAA Chain AAA, MgGH51 [Meripilus giganteus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P82593 | 4.25e-64 | 239 | 694 | 36 | 563 | Extracellular exo-alpha-L-arabinofuranosidase OS=Streptomyces chartreusis OX=1969 PE=1 SV=1 |
| Q9SG80 | 5.47e-35 | 239 | 706 | 42 | 576 | Alpha-L-arabinofuranosidase 1 OS=Arabidopsis thaliana OX=3702 GN=ASD1 PE=1 SV=1 |
| Q8VZR2 | 1.27e-34 | 231 | 680 | 33 | 551 | Alpha-L-arabinofuranosidase 2 OS=Arabidopsis thaliana OX=3702 GN=ASD2 PE=2 SV=1 |
| U6A629 | 3.19e-22 | 264 | 641 | 43 | 478 | Alpha-L-arabinofuranosidase A OS=Penicillium canescens OX=5083 GN=abfA PE=1 SV=1 |
| Q2U790 | 2.13e-16 | 239 | 720 | 27 | 575 | Probable alpha-L-arabinofuranosidase A OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=abfA PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
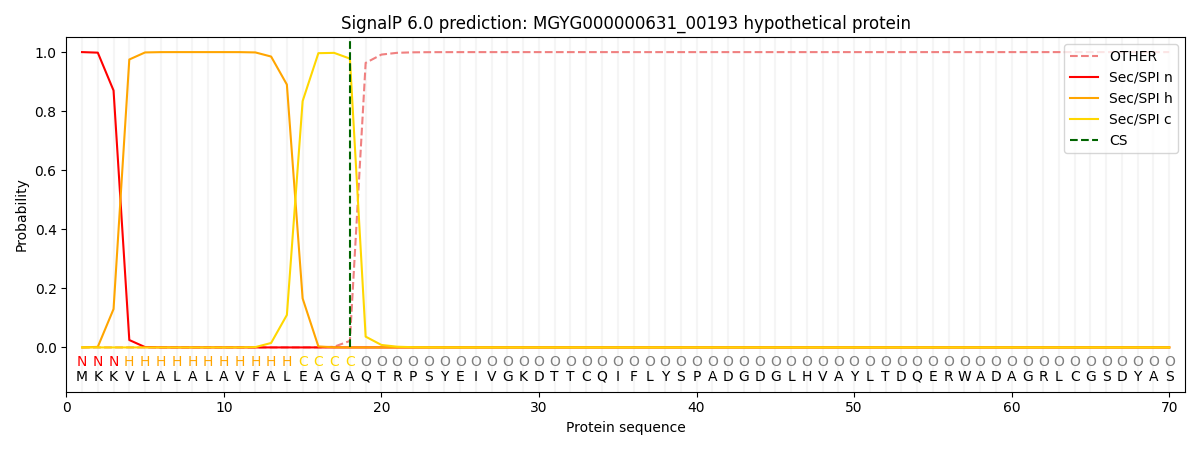
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000258 | 0.999109 | 0.000172 | 0.000157 | 0.000156 | 0.000141 |
