You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000633_00783
Basic Information
help
| Species |
Victivallis sp900768515
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900768515
|
| CAZyme ID |
MGYG000000633_00783
|
| CAZy Family |
GH148 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1290 |
|
143134.04 |
7.4248 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000633 |
2845403 |
MAG |
Madagascar |
Africa |
|
| Gene Location |
Start: 19138;
End: 23010
Strand: +
|
No EC number prediction in MGYG000000633_00783.
| Family |
Start |
End |
Evalue |
family coverage |
| GH148 |
676 |
830 |
1.2e-36 |
0.993421052631579 |
| GH148 |
889 |
1032 |
1.4e-25 |
0.9342105263157895 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| PRK03757
|
PRK03757 |
0.007 |
1 |
51 |
1 |
47 |
YceI family protein. |
This protein is predicted as SP
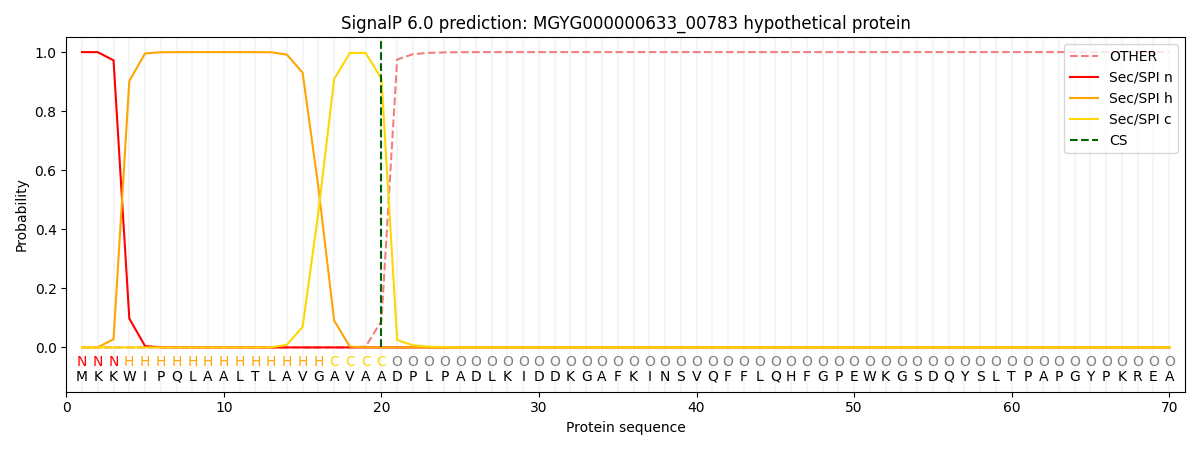
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000235
|
0.999156
|
0.000159
|
0.000163
|
0.000154
|
0.000138
|
There is no transmembrane helices in MGYG000000633_00783.

