You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000633_00833
You are here: Home > Sequence: MGYG000000633_00833
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900768515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900768515 | |||||||||||
| CAZyme ID | MGYG000000633_00833 | |||||||||||
| CAZy Family | CBM85 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 95611; End: 97395 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 244 | 530 | 1e-59 | 0.9504950495049505 |
| CBM85 | 42 | 178 | 4.1e-22 | 0.9621212121212122 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.69e-43 | 286 | 533 | 1 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 2.11e-39 | 248 | 528 | 11 | 303 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.33e-33 | 274 | 528 | 55 | 330 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCW34445.1 | 9.68e-121 | 42 | 593 | 52 | 588 |
| QDU72038.1 | 3.35e-116 | 83 | 593 | 107 | 605 |
| BAM03107.1 | 7.29e-109 | 83 | 593 | 99 | 606 |
| ACB76489.1 | 2.60e-104 | 66 | 585 | 80 | 596 |
| ATC63818.1 | 8.63e-104 | 44 | 593 | 272 | 817 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7D88_A | 1.69e-56 | 200 | 568 | 39 | 397 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 7D89_A | 1.23e-55 | 200 | 568 | 39 | 397 | ChainA, Beta-xylanase [Bacillus sp. (in: Bacteria)] |
| 1XYZ_A | 2.87e-22 | 250 | 535 | 40 | 342 | ChainA, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus],1XYZ_B Chain B, 1,4-BETA-D-XYLAN-XYLANOHYDROLASE [Acetivibrio thermocellus] |
| 3WUF_A | 1.55e-20 | 258 | 529 | 19 | 304 | Themutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 [Streptomyces sp.],3WUG_A The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
| 3WUB_A | 5.16e-20 | 258 | 529 | 19 | 304 | Thewild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 [Streptomyces sp.],3WUE_A The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 [Streptomyces sp.] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| A3DH97 | 2.62e-56 | 205 | 574 | 380 | 739 | Anti-sigma-I factor RsgI6 OS=Acetivibrio thermocellus (strain ATCC 27405 / DSM 1237 / JCM 9322 / NBRC 103400 / NCIMB 10682 / NRRL B-4536 / VPI 7372) OX=203119 GN=rsgI6 PE=1 SV=1 |
| A0A1P8AWH8 | 8.97e-47 | 210 | 593 | 565 | 940 | Endo-1,4-beta-xylanase 1 OS=Arabidopsis thaliana OX=3702 GN=XYN1 PE=1 SV=1 |
| O80596 | 1.46e-42 | 212 | 576 | 689 | 1045 | Endo-1,4-beta-xylanase 2 OS=Arabidopsis thaliana OX=3702 GN=XYN2 PE=3 SV=1 |
| F4JG10 | 1.01e-40 | 210 | 593 | 370 | 747 | Endo-1,4-beta-xylanase 3 OS=Arabidopsis thaliana OX=3702 GN=XYN3 PE=2 SV=1 |
| Q680B7 | 2.84e-28 | 145 | 583 | 91 | 553 | Endo-1,4-beta-xylanase 4 OS=Arabidopsis thaliana OX=3702 GN=XYN4 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
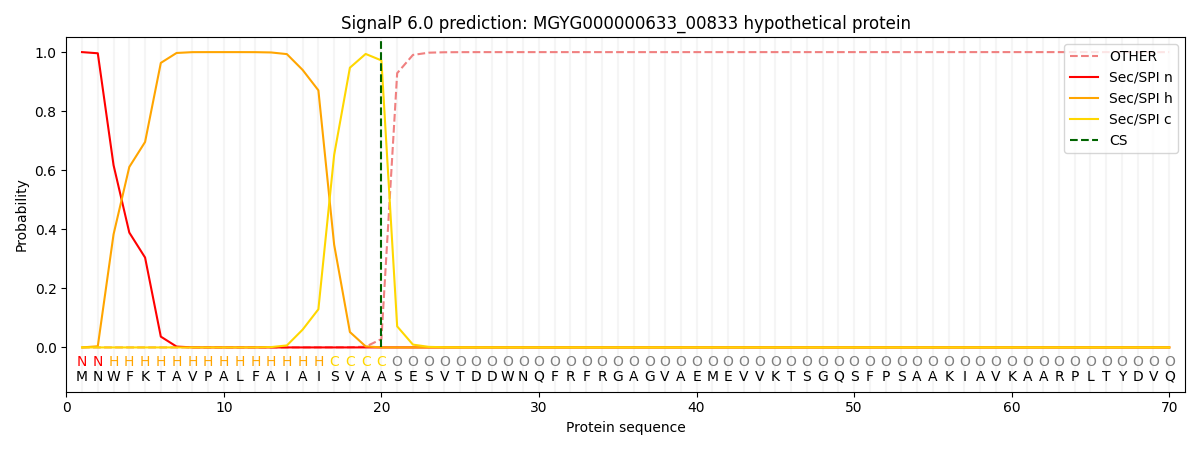
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000610 | 0.998539 | 0.000276 | 0.000181 | 0.000180 | 0.000178 |
