You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000633_00928
You are here: Home > Sequence: MGYG000000633_00928
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Victivallis sp900768515 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; Victivallis sp900768515 | |||||||||||
| CAZyme ID | MGYG000000633_00928 | |||||||||||
| CAZy Family | CE7 | |||||||||||
| CAZyme Description | Acetyl esterase Axe7A | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 780; End: 2060 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE7 | 136 | 419 | 4.2e-59 | 0.9456869009584664 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05448 | AXE1 | 8.98e-33 | 142 | 409 | 18 | 299 | Acetyl xylan esterase (AXE1). This family consists of several bacterial acetyl xylan esterase proteins. Acetyl xylan esterases are enzymes that hydrolyze the ester linkages of the acetyl groups in position 2 and/or 3 of the xylose moieties of natural acetylated xylan from hardwood. These enzymes are one of the accessory enzymes which are part of the xylanolytic system, together with xylanases, beta-xylosidases, alpha-arabinofuranosidases and methylglucuronidases; these are all required for the complete hydrolysis of xylan. |
| COG3458 | Axe1 | 3.14e-31 | 119 | 409 | 1 | 299 | Cephalosporin-C deacetylase or related acetyl esterase [Secondary metabolites biosynthesis, transport and catabolism]. |
| COG1506 | DAP2 | 1.15e-07 | 193 | 420 | 378 | 605 | Dipeptidyl aminopeptidase/acylaminoacyl peptidase [Amino acid transport and metabolism]. |
| COG0412 | DLH | 0.001 | 288 | 424 | 100 | 216 | Dienelactone hydrolase [Secondary metabolites biosynthesis, transport and catabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46416.1 | 9.17e-128 | 12 | 411 | 14 | 402 |
| SDU24469.1 | 9.80e-128 | 19 | 411 | 18 | 402 |
| AVM45055.1 | 3.37e-107 | 26 | 409 | 260 | 643 |
| AVM45740.1 | 1.37e-106 | 26 | 409 | 176 | 552 |
| AHF91279.1 | 5.97e-103 | 12 | 409 | 15 | 417 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1ODS_A | 1.69e-18 | 119 | 409 | 1 | 298 | CephalosporinC deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_B Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_C Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_D Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_E Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_F Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_G Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis],1ODS_H Cephalosporin C deacetylase from Bacillus subtilis [Bacillus subtilis] |
| 1L7A_A | 1.69e-18 | 119 | 409 | 1 | 298 | structuralGenomics, crystal structure of Cephalosporin C deacetylase [Bacillus subtilis],1L7A_B structural Genomics, crystal structure of Cephalosporin C deacetylase [Bacillus subtilis] |
| 1ODT_C | 3.11e-18 | 119 | 409 | 1 | 298 | cephalosporinC deacetylase mutated, in complex with acetate [Bacillus subtilis],1ODT_H cephalosporin C deacetylase mutated, in complex with acetate [Bacillus subtilis] |
| 6AGQ_A | 8.98e-17 | 119 | 409 | 1 | 303 | Acetylxylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_B Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_C Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_D Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_E Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4],6AGQ_F Acetyl xylan esterase from Paenibacillus sp. R4 [Paenibacillus sp. R4] |
| 5GMA_A | 1.43e-16 | 119 | 409 | 13 | 315 | Crystalstructure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_B Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_C Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_D Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_E Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8],5GMA_F Crystal structure of the P228A variant of Thermotoga maritima acetyl esterase [Thermotoga maritima MSB8] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EXI2 | 1.25e-33 | 33 | 398 | 52 | 409 | Acetyl esterase Axe7A OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=axe7A PE=1 SV=1 |
| P94388 | 6.83e-18 | 119 | 409 | 1 | 298 | Cephalosporin-C deacetylase OS=Bacillus subtilis (strain 168) OX=224308 GN=cah PE=1 SV=1 |
| Q9WXT2 | 6.94e-16 | 119 | 409 | 1 | 303 | Cephalosporin-C deacetylase OS=Thermotoga maritima (strain ATCC 43589 / DSM 3109 / JCM 10099 / NBRC 100826 / MSB8) OX=243274 GN=axeA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
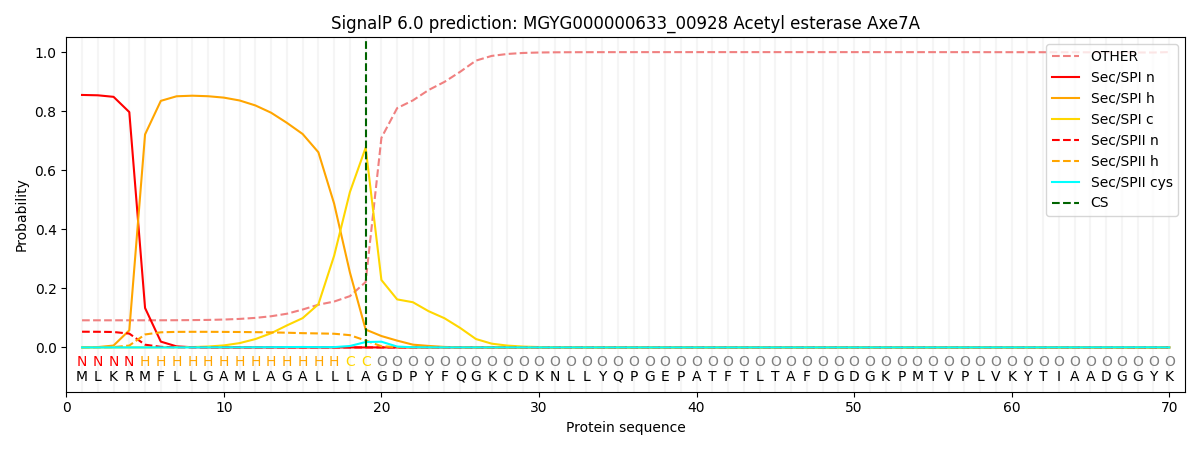
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.099830 | 0.843410 | 0.055600 | 0.000455 | 0.000324 | 0.000347 |
