You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000644_01606
You are here: Home > Sequence: MGYG000000644_01606
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp900546535 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp900546535 | |||||||||||
| CAZyme ID | MGYG000000644_01606 | |||||||||||
| CAZy Family | CE8 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 12061; End: 15249 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE8 | 655 | 893 | 5.5e-48 | 0.8402777777777778 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4677 | PemB | 1.32e-19 | 662 | 914 | 112 | 397 | Pectin methylesterase and related acyl-CoA thioesterases [Carbohydrate transport and metabolism, Lipid transport and metabolism]. |
| PLN02432 | PLN02432 | 6.45e-18 | 662 | 852 | 40 | 222 | putative pectinesterase |
| PRK10531 | PRK10531 | 7.87e-14 | 707 | 887 | 194 | 416 | putative acyl-CoA thioester hydrolase. |
| PLN02773 | PLN02773 | 1.43e-13 | 662 | 885 | 34 | 263 | pectinesterase |
| pfam01095 | Pectinesterase | 2.95e-12 | 662 | 844 | 29 | 220 | Pectinesterase. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VEH16322.1 | 2.08e-210 | 157 | 1062 | 562 | 1471 |
| AGB28243.1 | 1.19e-187 | 155 | 1062 | 558 | 1371 |
| QUT74167.1 | 2.55e-115 | 22 | 1062 | 262 | 1435 |
| QCD38476.1 | 3.14e-88 | 644 | 1062 | 1049 | 1476 |
| QCP72166.1 | 3.14e-88 | 644 | 1062 | 1049 | 1476 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 1XG2_A | 2.04e-08 | 622 | 807 | 2 | 175 | ChainA, Pectinesterase 1 [Solanum lycopersicum] |
| 4PMH_A | 2.69e-08 | 701 | 844 | 139 | 292 | Thestructure of rice weevil pectin methyl esterase [Sitophilus oryzae] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9SKX2 | 4.55e-12 | 657 | 885 | 237 | 463 | Probable pectinesterase/pectinesterase inhibitor 16 OS=Arabidopsis thaliana OX=3702 GN=PME16 PE=2 SV=1 |
| Q9ZQA3 | 2.03e-10 | 659 | 858 | 115 | 321 | Probable pectinesterase 15 OS=Arabidopsis thaliana OX=3702 GN=PME15 PE=2 SV=1 |
| Q9LYT5 | 9.04e-10 | 660 | 883 | 253 | 471 | Probable pectinesterase/pectinesterase inhibitor 35 OS=Arabidopsis thaliana OX=3702 GN=PME35 PE=2 SV=1 |
| Q9LUL8 | 6.81e-09 | 665 | 857 | 691 | 887 | Putative pectinesterase/pectinesterase inhibitor 26 OS=Arabidopsis thaliana OX=3702 GN=PME26 PE=2 SV=1 |
| O64479 | 4.11e-08 | 665 | 860 | 71 | 270 | Putative pectinesterase 10 OS=Arabidopsis thaliana OX=3702 GN=PME10 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
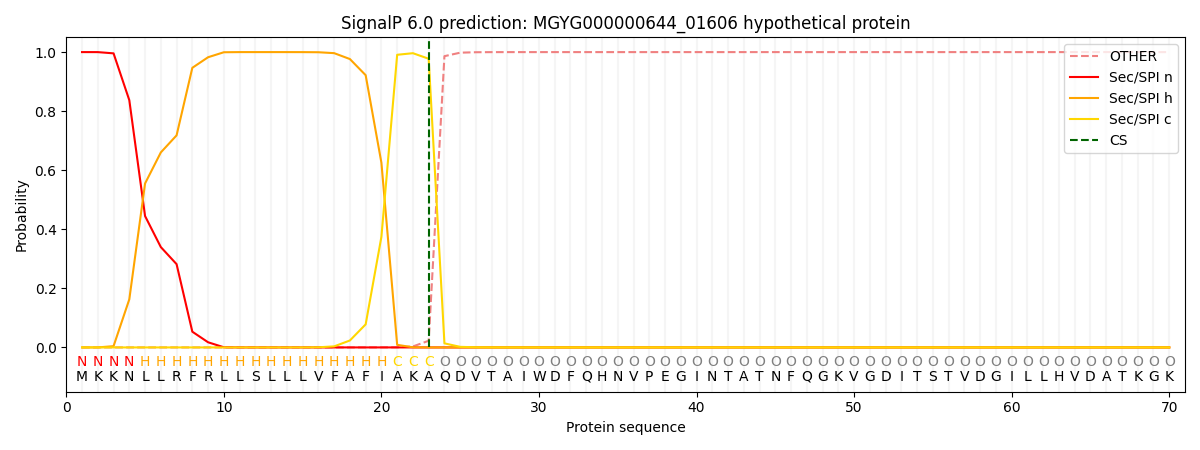
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000235 | 0.999081 | 0.000211 | 0.000164 | 0.000160 | 0.000152 |
