You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000652_02297
You are here: Home > Sequence: MGYG000000652_02297
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides togonis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides togonis | |||||||||||
| CAZyme ID | MGYG000000652_02297 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 25179; End: 26036 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 136 | 279 | 2e-17 | 0.9274193548387096 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 3.04e-14 | 130 | 277 | 1 | 125 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd00057 | FA58C | 2.22e-07 | 135 | 264 | 15 | 141 | Substituted updates: Jan 31, 2002 |
| cd14948 | BACON | 7.84e-04 | 69 | 115 | 34 | 83 | Bacteroidetes-Associated Carbohydrate-binding (putative) Often N-terminal (BACON) domain. The BACON domain is found in diverse domain architectures and accociated with a wide variety of domains, including carbohydrate-active enzymes and proteases. It was named for its suggested function of carbohydrate binding; the latter was inferred from domain architectures, sequence conservation, and phyletic distribution. However, recent experimental data suggest that its primary function in Bacteroides ovatus endo-xyloglucanase BoGH5A is to distance the catalytic module from the cell surface and confer additional mobility to the catalytic domain for attack of the polysaccharide. No evidence for a direct role in carbohydrate binding could be found in that case. The large majority of BACON domains are found in Bacteroidetes. |
| smart00231 | FA58C | 0.004 | 154 | 264 | 34 | 136 | Coagulation factor 5/8 C-terminal domain, discoidin domain. Cell surface-attached carbohydrate-binding domain, present in eukaryotes and assumed to have horizontally transferred to eubacterial genomes. |
| pfam13004 | BACON | 0.005 | 61 | 115 | 1 | 61 | Putative binding domain, N-terminal. The BACON (Bacteroidetes-Associated Carbohydrate-binding Often N-terminal) domain is an all-beta domain found in diverse architectures, principally in combination with carbohydrate-active enzymes and proteases. These architectures suggest a carbohydrate-binding function which is also supported by the nature of BACON's few conserved amino-acids. The phyletic distribution of BACON and other data tentatively suggest that it may frequently function to bind mucin. Further work with the characterized structure of a member of glycoside hydrolase family 5 enzyme, Structure 3ZMR, has found no evidence for carbohydrate-binding for this domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| CCO21431.1 | 3.47e-117 | 1 | 285 | 5 | 286 |
| QIL39112.1 | 8.47e-11 | 115 | 266 | 172 | 301 |
| AZU64122.1 | 1.15e-09 | 108 | 283 | 1198 | 1357 |
| AIM35717.1 | 1.43e-09 | 125 | 282 | 317 | 454 |
| BCA48665.1 | 1.69e-09 | 120 | 273 | 242 | 380 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2KD7_A | 2.47e-11 | 135 | 224 | 21 | 106 | ChainA, Putative chitobiase [Bacteroides thetaiotaomicron VPI-5482] |
| 3GGL_A | 1.98e-10 | 135 | 224 | 31 | 116 | X-RayStructure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. [Bacteroides thetaiotaomicron],6OE2_A X-Ray Structure of the C-terminal domain (277-440) of Putative chitobiase from Bacteroides thetaiotaomicron. Northeast Structural Genomics Consortium Target BtR324A. Re-refinement of 3GGL with correct metal Mn replacing Zn. New metal confirmed with PIXE analysis of original sample. [Bacteroides thetaiotaomicron] |
| 3F2Z_A | 7.99e-10 | 118 | 224 | 2 | 106 | Crystalstructure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B [Bacteroides fragilis NCTC 9343] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
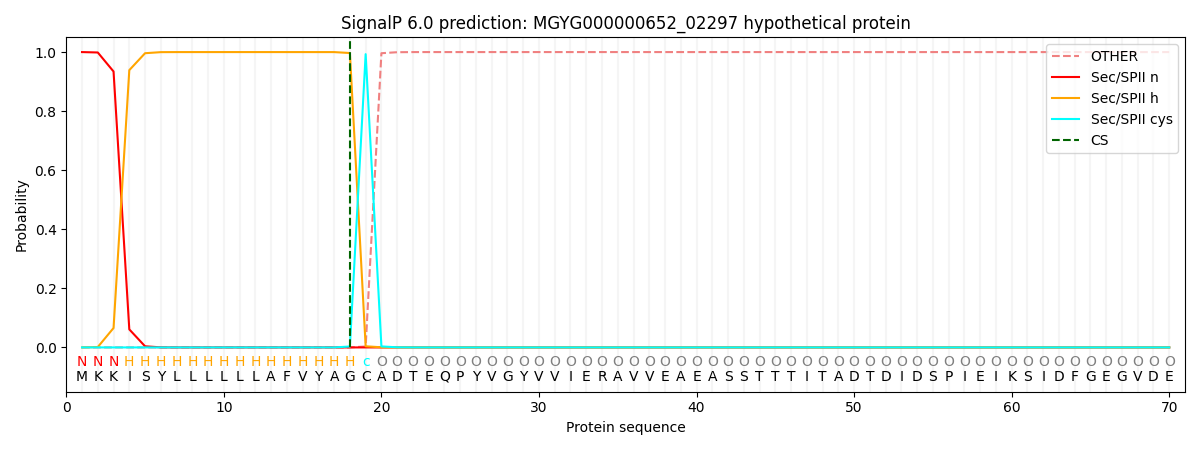
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000073 | 0.000000 | 0.000000 | 0.000000 |
