You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000653_01761
You are here: Home > Sequence: MGYG000000653_01761
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA11471 sp000434215 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA11471; UBA11471; UBA11471 sp000434215 | |||||||||||
| CAZyme ID | MGYG000000653_01761 | |||||||||||
| CAZy Family | PL1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 18531; End: 21404 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| PL1 | 77 | 261 | 1.3e-49 | 0.994535519125683 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13385 | Laminin_G_3 | 4.39e-19 | 697 | 859 | 3 | 151 | Concanavalin A-like lectin/glucanases superfamily. This domain belongs to the Concanavalin A-like lectin/glucanases superfamily. |
| TIGR04183 | Por_Secre_tail | 1.49e-09 | 882 | 956 | 1 | 71 | Por secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber average twenty or more copies of a C-terminal domain, represented by this model, associated with sorting to the outer membrane and covalent modification. |
| smart00560 | LamGL | 7.34e-09 | 761 | 854 | 37 | 131 | LamG-like jellyroll fold domain. |
| pfam18962 | Por_Secre_tail | 5.05e-07 | 882 | 951 | 1 | 67 | Secretion system C-terminal sorting domain. Species that include Porphyromonas gingivalis, Fibrobacter succinogenes, Flavobacterium johnsoniae, Cytophaga hutchinsonii, Gramella forsetii, Prevotella intermedia, and Salinibacter ruber have on average twenty or more copies of this C-terminal domain, associated with sorting to the outer membrane and covalent modification. This domain targets proteins to type IX secretion systems and is secreted then cleaved off by a C-terminal signal peptidease. Based on similarity to other families it is likely that this domain adopts an immunoglobulin like fold. |
| smart00282 | LamG | 6.20e-06 | 745 | 850 | 19 | 131 | Laminin G domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT73817.1 | 1.64e-272 | 14 | 956 | 14 | 972 |
| QDH80543.1 | 2.23e-181 | 10 | 931 | 18 | 914 |
| AGA79623.1 | 1.16e-180 | 22 | 931 | 33 | 914 |
| ANF52864.1 | 7.53e-173 | 22 | 947 | 30 | 989 |
| AYN02029.1 | 2.53e-170 | 16 | 897 | 24 | 938 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B8NQQ7 | 7.27e-110 | 21 | 447 | 18 | 416 | Probable pectate lyase C OS=Aspergillus flavus (strain ATCC 200026 / FGSC A1120 / IAM 13836 / NRRL 3357 / JCM 12722 / SRRC 167) OX=332952 GN=plyC PE=3 SV=1 |
| Q0CLG7 | 1.51e-108 | 21 | 447 | 18 | 416 | Probable pectate lyase C OS=Aspergillus terreus (strain NIH 2624 / FGSC A1156) OX=341663 GN=plyC PE=3 SV=1 |
| A1DPF0 | 4.28e-108 | 23 | 447 | 21 | 417 | Probable pectate lyase C OS=Neosartorya fischeri (strain ATCC 1020 / DSM 3700 / CBS 544.65 / FGSC A1164 / JCM 1740 / NRRL 181 / WB 181) OX=331117 GN=plyC PE=3 SV=1 |
| Q2UB83 | 1.14e-107 | 21 | 447 | 18 | 416 | Probable pectate lyase C OS=Aspergillus oryzae (strain ATCC 42149 / RIB 40) OX=510516 GN=plyC PE=3 SV=1 |
| Q4WL88 | 3.23e-107 | 23 | 447 | 21 | 417 | Probable pectate lyase C OS=Neosartorya fumigata (strain ATCC MYA-4609 / Af293 / CBS 101355 / FGSC A1100) OX=330879 GN=plyC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
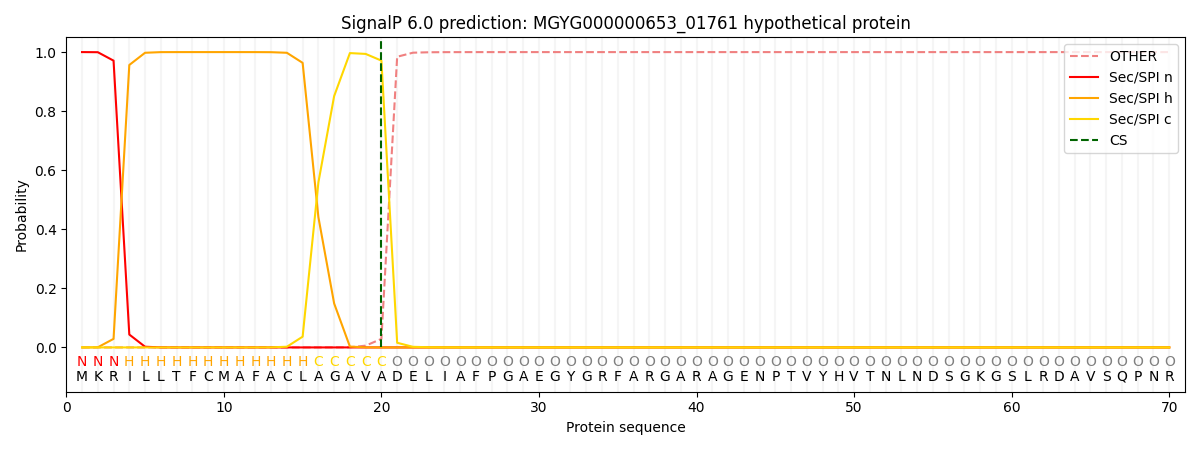
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.999080 | 0.000203 | 0.000150 | 0.000146 | 0.000142 |
