You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000655_02118
You are here: Home > Sequence: MGYG000000655_02118
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
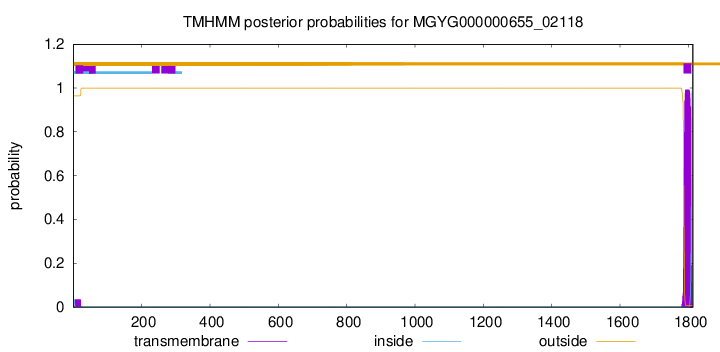
TMHMM annotations
Basic Information help
| Species | Holdemanella sp900547815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Holdemanella; Holdemanella sp900547815 | |||||||||||
| CAZyme ID | MGYG000000655_02118 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 268; End: 5709 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 123 | 256 | 1.1e-16 | 0.8467741935483871 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 1.98e-08 | 123 | 247 | 12 | 119 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 1.14e-07 | 1402 | 1520 | 9 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| pfam00754 | F5_F8_type_C | 7.58e-07 | 273 | 415 | 1 | 114 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| NF033777 | M_group_A_cterm | 1.39e-06 | 1608 | 1811 | 28 | 216 | M protein C-terminal domain. M protein (emm) is an important virulence protein and serology-defining surface antigen of Streptococcus pyogenes (group A Streptococcus). M protein has an amino-terminal YSIRK-type signal sequence (associated with cross-wall targeting in dividing cells), and a C-terminal LPXTG domain for processing by sortase and covalent attachment to the Gram-positive cell wall. Past the signal peptide, M protein has a hypervariable region, but this HMM describes only the well-conserved region C-terminal to the hypervariable region. It discriminates M protein from two related proteins, Enn and Mrp. |
| cd00057 | FA58C | 3.65e-05 | 273 | 416 | 13 | 131 | Substituted updates: Jan 31, 2002 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT17581.1 | 0.0 | 25 | 1608 | 41 | 1458 |
| ABG83191.1 | 0.0 | 2 | 1697 | 8 | 1544 |
| AOY54034.1 | 0.0 | 2 | 1670 | 8 | 1495 |
| ATD48595.1 | 0.0 | 2 | 1670 | 8 | 1495 |
| BAB80970.1 | 0.0 | 2 | 1697 | 8 | 1544 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4A41_A | 4.65e-09 | 1389 | 1521 | 32 | 158 | CpGH89CBM32-5,from Clostridium perfringens, in complex with galactose [Clostridium perfringens],4A44_A CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen [Clostridium perfringens],4A45_A CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose [Clostridium perfringens],4AAX_A CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DTR4 | 2.65e-10 | 1349 | 1521 | 473 | 642 | A type blood N-acetyl-alpha-D-galactosamine deacetylase OS=Flavonifractor plautii OX=292800 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
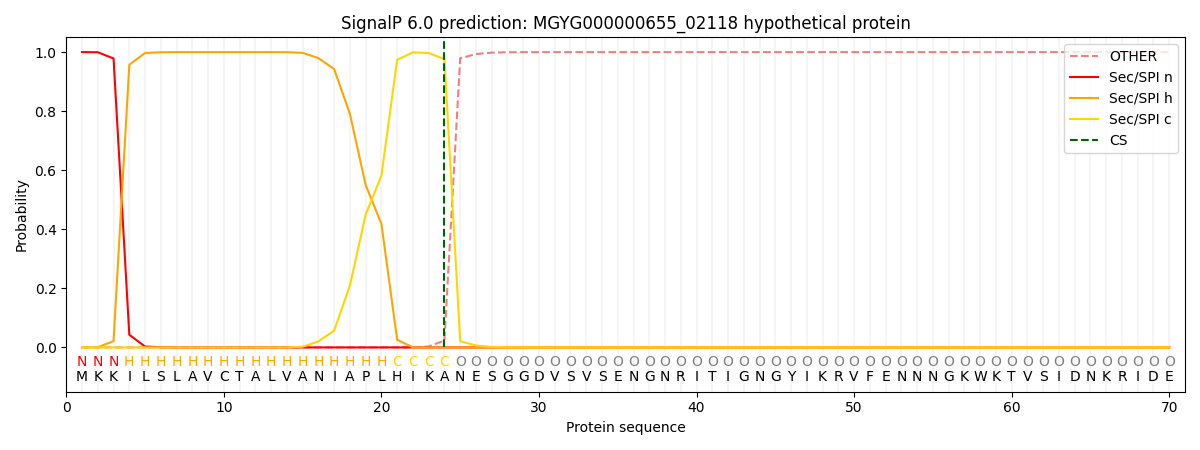
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000262 | 0.999026 | 0.000196 | 0.000170 | 0.000174 | 0.000153 |