You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000662_00244
You are here: Home > Sequence: MGYG000000662_00244
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
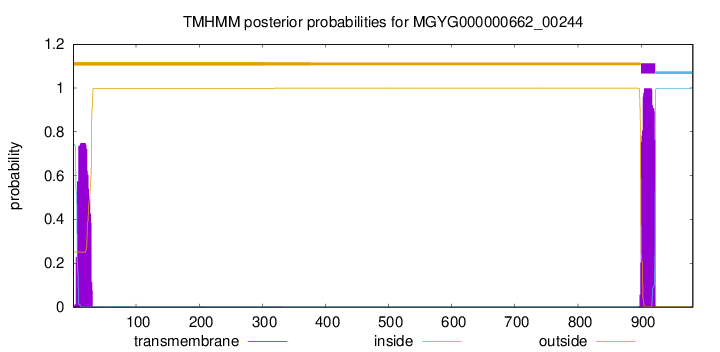
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Erysipelotrichales; Erysipelotrichaceae; Bulleidia; | |||||||||||
| CAZyme ID | MGYG000000662_00244 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 166; End: 3114 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4193 | LytD | 1.49e-12 | 235 | 425 | 59 | 237 | Beta- N-acetylglucosaminidase [Carbohydrate transport and metabolism]. |
| pfam01832 | Glucosaminidase | 2.27e-04 | 301 | 366 | 2 | 77 | Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase. This family includes Mannosyl-glycoprotein endo-beta-N-acetylglucosaminidase EC:3.2.1.96. As well as the flageller protein J that has been shown to hydrolyze peptidoglycan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QSI25075.1 | 2.20e-183 | 38 | 926 | 32 | 912 |
| QJA03888.1 | 1.33e-182 | 38 | 911 | 32 | 898 |
| QIX10256.1 | 9.60e-179 | 38 | 889 | 32 | 876 |
| QQR25465.1 | 1.35e-178 | 38 | 889 | 32 | 876 |
| ANU70664.1 | 1.35e-178 | 38 | 889 | 32 | 876 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4Q2W_A | 1.90e-15 | 169 | 424 | 41 | 290 | CrystalStructure of pneumococcal peptidoglycan hydrolase LytB [Streptococcus pneumoniae TIGR4] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P59205 | 1.80e-15 | 121 | 424 | 371 | 658 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae serotype 4 (strain ATCC BAA-334 / TIGR4) OX=170187 GN=lytB PE=1 SV=1 |
| P59206 | 2.55e-15 | 121 | 424 | 415 | 702 | Putative endo-beta-N-acetylglucosaminidase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=lytB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
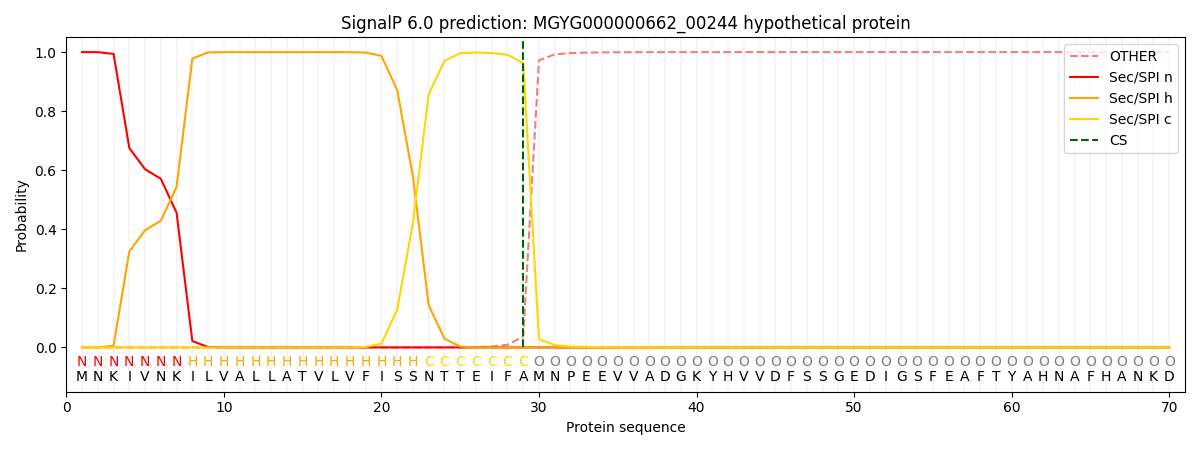
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000271 | 0.999053 | 0.000189 | 0.000159 | 0.000148 | 0.000137 |