You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000666_00020
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; ;
|
| CAZyme ID |
MGYG000000666_00020
|
| CAZy Family |
GH148 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 1103 |
|
124181.14 |
9.8017 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000666 |
4523435 |
MAG |
Kazakhstan |
Asia |
|
| Gene Location |
Start: 25258;
End: 28569
Strand: -
|
No EC number prediction in MGYG000000666_00020.
| Family |
Start |
End |
Evalue |
family coverage |
| GH148 |
473 |
619 |
1.4e-35 |
0.993421052631579 |
| GH148 |
675 |
795 |
1.4e-19 |
0.7763157894736842 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG2730
|
BglC |
5.50e-04 |
499 |
619 |
82 |
190 |
Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
This protein is predicted as SP
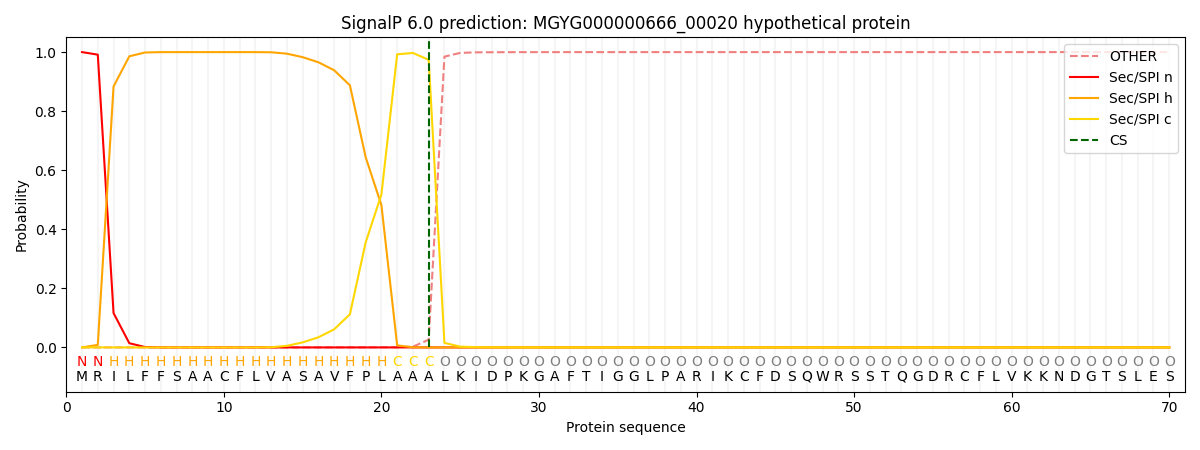
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000225
|
0.999184
|
0.000154
|
0.000145
|
0.000136
|
0.000126
|