You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000666_00078
You are here: Home > Sequence: MGYG000000666_00078
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; UBA1829; ; | |||||||||||
| CAZyme ID | MGYG000000666_00078 | |||||||||||
| CAZy Family | GH71 | |||||||||||
| CAZyme Description | Hercynine oxygenase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 51356; End: 54037 Strand: + | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03659 | Glyco_hydro_71 | 2.17e-49 | 26 | 420 | 1 | 372 | Glycosyl hydrolase family 71. Family of alpha-1,3-glucanases. |
| pfam03781 | FGE-sulfatase | 2.81e-47 | 608 | 879 | 1 | 258 | Sulfatase-modifying factor enzyme 1. This domain is found in eukaryotic proteins required for post-translational sulfatase modification (SUMF1). These proteins are associated with the rare disorder multiple sulfatase deficiency (MSD). The protein product of the SUMF1 gene is FGE, formylglycine (FGly),-generating enzyme, which is a sulfatase. Sulfatases are enzymes essential for degradation and remodelling of sulfate esters, and formylglycine (FGly), the key catalytic in the active site, is unique to sulfatases. FGE is localized to the endoplasmic reticulum (ER) and interacts with and modifies the unfolded form of newly synthesized sulfatases. FGE is a single-domain monomer with a surprising paucity of secondary structure that adopts a unique fold which is stabilized by two Ca2+ ions. The effect of all mutations found in MSD patients is explained by the FGE structure, providing a molecular basis for MSD. A redox-active disulfide bond is present in the active site of FGE. An oxidized cysteine residue, possibly cysteine sulfenic acid, has been detected that may allow formulation of a structure-based mechanism for FGly formation from cysteine residues in all sulfatases. In Mycobacteria and Treponema denticola this enzyme functions as an iron(II)-dependent oxidoreductase. |
| COG1262 | YfmG | 1.71e-42 | 600 | 879 | 41 | 310 | Formylglycine-generating enzyme, required for sulfatase activity, contains SUMF1/FGE domain [Posttranslational modification, protein turnover, chaperones]. |
| cd11577 | GH71 | 2.15e-39 | 20 | 316 | 1 | 272 | Glycoside hydrolase family 71. This family of glycoside hydrolases 71 (following the CAZY nomenclature) function as alpha-1,3-glucanases (mutanases, EC 3.2.1.59). They appear to have an endo-hydrolytic mode of enzymatic activity and bacterial members are investigated as candidates for the development of dental caries treatments.The member from fission yeast, endo-alpha-1,3-glucanase Agn1p, plays a vital role in daughter cell separation, while Agn2p has been associated with endolysis of the ascus wall. |
| TIGR03529 | GldK_short | 7.33e-22 | 607 | 880 | 48 | 336 | gliding motility-associated lipoprotein GldK. Members of this protein family are exclusive to the Bacteroidetes phylum (previously Cytophaga-Flavobacteria-Bacteroides). GldK is a lipoprotein linked to a type of rapid surface gliding motility found in certain Bacteroidetes, such as Flavobacterium johnsoniae. Knockouts of GldK abolish the gliding phenotype. GldK is homologous to GldJ. This model represents a GldK homolog in Cytophaga hutchinsonii and several other species that has a different, shorter architecture than that found in Flavobacterium johnsoniae and related species (represented by (TIGR03525). Gliding motility appears closely linked to chitin utilization in the model species Flavobacterium johnsoniae. Bacteroidetes with members of this protein family appear to have all of the genes associated with gliding motility. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM43180.1 | 4.94e-247 | 12 | 887 | 13 | 835 |
| BCF90409.1 | 1.85e-40 | 24 | 427 | 9 | 406 |
| AXE95223.1 | 5.15e-36 | 9 | 427 | 19 | 432 |
| AUT54417.1 | 3.85e-35 | 24 | 440 | 30 | 440 |
| ALP64444.1 | 3.85e-35 | 24 | 440 | 30 | 440 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7ML6_A | 1.00e-19 | 601 | 889 | 10 | 313 | ChainA, CalU17 [Micromonospora echinospora],7MSY_A Chain A, CalU17 [Micromonospora echinospora] |
| 6VQP_A | 1.31e-19 | 601 | 889 | 28 | 331 | ChainA, CalU17 [Micromonospora echinospora] |
| 6MUJ_A | 1.88e-19 | 613 | 866 | 32 | 295 | Formylglycinegenerating enzyme bound to copper [Streptomyces coelicolor A3(2)],6MUJ_B Formylglycine generating enzyme bound to copper [Streptomyces coelicolor A3(2)],6MUJ_C Formylglycine generating enzyme bound to copper [Streptomyces coelicolor A3(2)],6MUJ_D Formylglycine generating enzyme bound to copper [Streptomyces coelicolor A3(2)],6MUJ_E Formylglycine generating enzyme bound to copper [Streptomyces coelicolor A3(2)] |
| 2Q17_A | 2.77e-19 | 613 | 866 | 59 | 322 | FormylglycineGenerating Enzyme from Streptomyces coelicolor [Streptomyces coelicolor A3(2)],2Q17_B Formylglycine Generating Enzyme from Streptomyces coelicolor [Streptomyces coelicolor A3(2)],2Q17_C Formylglycine Generating Enzyme from Streptomyces coelicolor [Streptomyces coelicolor A3(2)],2Q17_D Formylglycine Generating Enzyme from Streptomyces coelicolor [Streptomyces coelicolor A3(2)],2Q17_E Formylglycine Generating Enzyme from Streptomyces coelicolor [Streptomyces coelicolor A3(2)] |
| 1Y1F_X | 1.80e-18 | 604 | 877 | 9 | 292 | humanformylglycine generating enzyme with cysteine sulfenic acid [Homo sapiens],1Y1J_X human formylglycine generating enzyme, sulfonic acid/desulfurated form [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O13716 | 1.01e-22 | 8 | 440 | 6 | 420 | Glucan endo-1,3-alpha-glucosidase agn1 OS=Schizosaccharomyces pombe (strain 972 / ATCC 24843) OX=284812 GN=agn1 PE=1 SV=2 |
| Q9F3C7 | 9.51e-19 | 613 | 866 | 27 | 290 | Formylglycine-generating enzyme OS=Streptomyces coelicolor (strain ATCC BAA-471 / A3(2) / M145) OX=100226 GN=SCO7548 PE=1 SV=1 |
| V9TSX0 | 1.32e-18 | 658 | 849 | 49 | 248 | Chaperone protein LppX OS=Paenibacillus barcinonensis OX=198119 GN=lppX PE=3 SV=1 |
| Q0P5L5 | 1.61e-18 | 602 | 884 | 79 | 374 | Formylglycine-generating enzyme OS=Bos taurus OX=9913 GN=SUMF1 PE=2 SV=1 |
| Q8R0F3 | 6.82e-18 | 601 | 877 | 78 | 362 | Formylglycine-generating enzyme OS=Mus musculus OX=10090 GN=Sumf1 PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
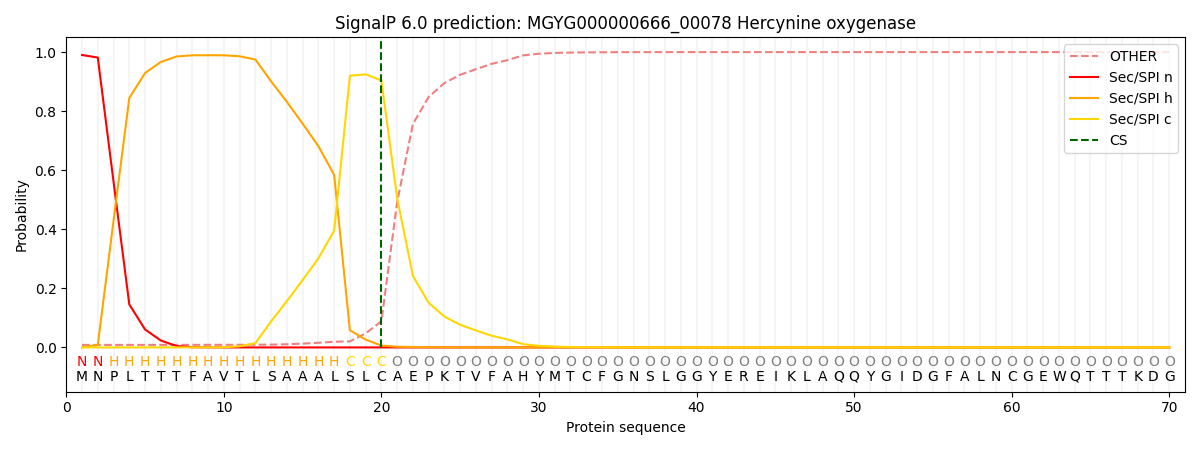
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.011345 | 0.985826 | 0.002054 | 0.000247 | 0.000240 | 0.000247 |
