You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000667_00487
Basic Information
help
| Species |
UBA7173 sp900546835
|
| Lineage |
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Muribaculaceae; UBA7173; UBA7173 sp900546835
|
| CAZyme ID |
MGYG000000667_00487
|
| CAZy Family |
GH147 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000667 |
2502097 |
MAG |
Kazakhstan |
Asia |
|
| Gene Location |
Start: 37065;
End: 39098
Strand: +
|
| Family |
Start |
End |
Evalue |
family coverage |
| GH147 |
22 |
674 |
1.6e-248 |
0.7769607843137255 |
MGYG000000667_00487 has no CDD domain.
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
| 4QP0_A
|
3.26e-07 |
85 |
204 |
59 |
176 |
CrystalStructure Analysis of the Endo-1,4-beta-mannanase from Rhizomucor miehei [Rhizomucor miehei] |
This protein is predicted as SP
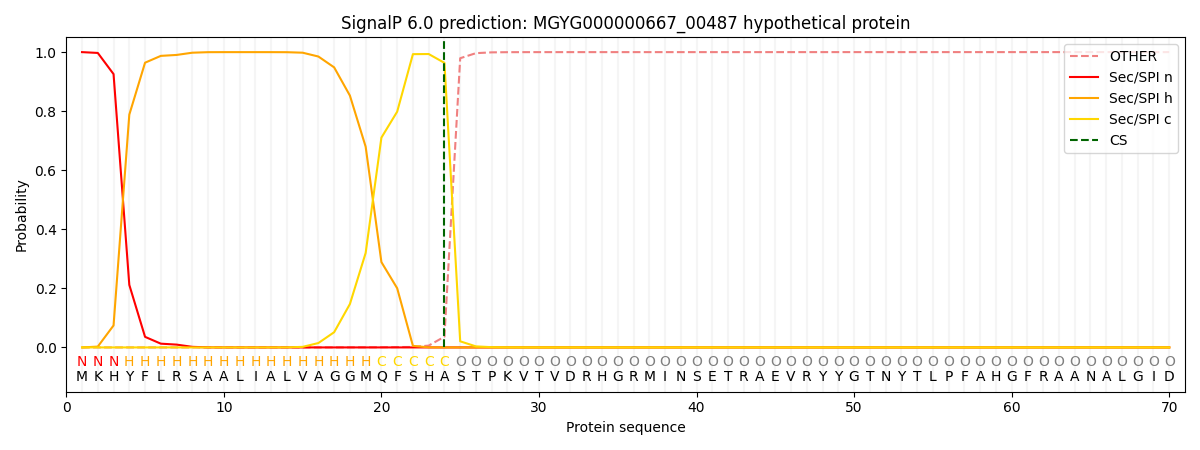
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.000404
|
0.998531
|
0.000423
|
0.000233
|
0.000200
|
0.000177
|
There is no transmembrane helices in MGYG000000667_00487.

