You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000672_00762
You are here: Home > Sequence: MGYG000000672_00762
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp000431015 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp000431015 | |||||||||||
| CAZyme ID | MGYG000000672_00762 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 135808; End: 139074 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 351 | 1029 | 2e-82 | 0.6848404255319149 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 8.32e-28 | 375 | 967 | 25 | 627 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 2.39e-11 | 437 | 789 | 125 | 449 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| pfam00754 | F5_F8_type_C | 2.06e-10 | 245 | 337 | 23 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| PRK10150 | PRK10150 | 4.49e-09 | 372 | 743 | 27 | 352 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 1.27e-06 | 551 | 664 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BAR52607.1 | 0.0 | 28 | 1074 | 72 | 1113 |
| AEW22509.1 | 0.0 | 28 | 1074 | 72 | 1113 |
| BAR49784.1 | 0.0 | 28 | 1074 | 72 | 1113 |
| ALO48125.1 | 0.0 | 26 | 1061 | 47 | 1073 |
| QRO23425.1 | 0.0 | 28 | 1074 | 64 | 1102 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5N6U_A | 4.78e-42 | 376 | 979 | 40 | 662 | Crystalstructure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_B Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_C Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12],5N6U_D Crystal structure of Beta-D-Mannosidase from Dictyoglomus thermophilum. [Dictyoglomus thermophilum H-6-12] |
| 6BYE_A | 7.65e-38 | 366 | 958 | 19 | 658 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306],6BYE_B Crystal structure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri in complex with mannose [Xanthomonas citri pv. citri str. 306] |
| 6BYC_A | 7.67e-38 | 366 | 958 | 19 | 658 | Crystalstructure of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYI_A | 1.76e-37 | 366 | 958 | 17 | 656 | Crystalstructure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYI_B Crystal structure of the acid-base mutant (E477A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
| 6BYG_A | 4.08e-37 | 366 | 958 | 19 | 658 | Crystalstructure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306],6BYG_B Crystal structure of the nucleophile mutant (E575A) of the GH2 exo-beta-mannanase from Xanthomonas axonopodis pv. citri [Xanthomonas citri pv. citri str. 306] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q75W54 | 1.27e-75 | 351 | 1059 | 10 | 780 | Mannosylglycoprotein endo-beta-mannosidase OS=Arabidopsis thaliana OX=3702 GN=EBM PE=1 SV=3 |
| Q5H7P5 | 5.67e-74 | 351 | 1020 | 7 | 742 | Mannosylglycoprotein endo-beta-mannosidase OS=Lilium longiflorum OX=4690 GN=EBM PE=1 SV=4 |
| Q82NR8 | 3.32e-63 | 354 | 1071 | 57 | 781 | Exo-beta-D-glucosaminidase OS=Streptomyces avermitilis (strain ATCC 31267 / DSM 46492 / JCM 5070 / NBRC 14893 / NCIMB 12804 / NRRL 8165 / MA-4680) OX=227882 GN=csxA PE=1 SV=1 |
| Q4R1C4 | 9.95e-40 | 353 | 1059 | 37 | 763 | Exo-beta-D-glucosaminidase OS=Hypocrea jecorina OX=51453 GN=gls93 PE=1 SV=1 |
| C0LRA7 | 7.01e-39 | 353 | 1059 | 35 | 761 | Exo-beta-D-glucosaminidase OS=Hypocrea virens OX=29875 GN=gls1 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
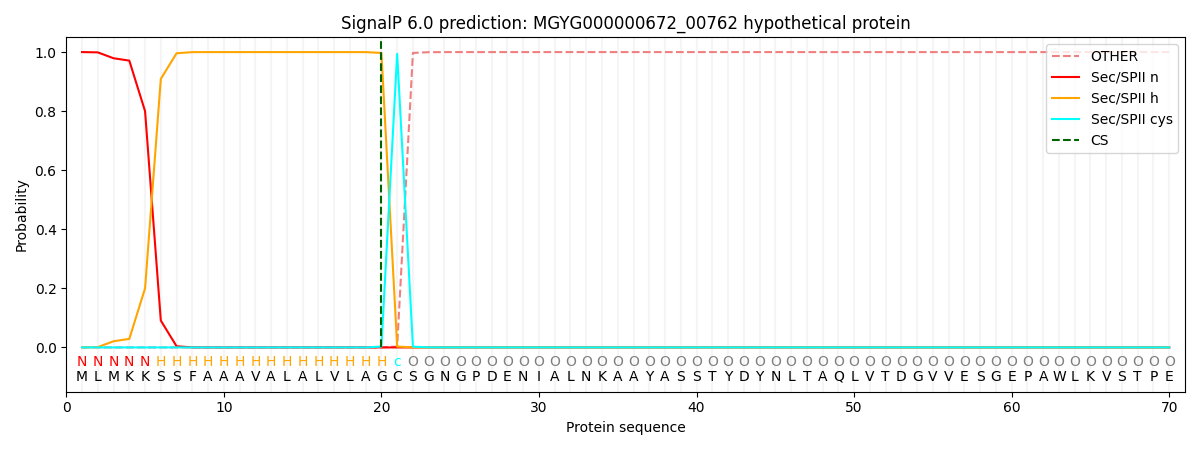
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000000 | 1.000040 | 0.000000 | 0.000000 | 0.000000 |
