You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000675_00831
You are here: Home > Sequence: MGYG000000675_00831
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
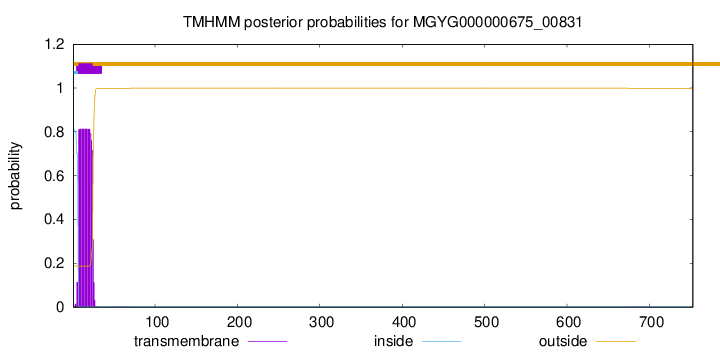
TMHMM annotations
Basic Information help
| Species | Bacteroides congonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides congonensis | |||||||||||
| CAZyme ID | MGYG000000675_00831 | |||||||||||
| CAZy Family | GH95 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 71508; End: 73769 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH95 | 112 | 689 | 1.1e-83 | 0.8365650969529086 |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQT78392.1 | 0.0 | 1 | 752 | 1 | 752 |
| QRP56363.1 | 0.0 | 1 | 752 | 1 | 752 |
| QUU06877.1 | 0.0 | 1 | 752 | 1 | 752 |
| ASM65081.1 | 0.0 | 1 | 752 | 1 | 752 |
| QDM12778.1 | 0.0 | 5 | 750 | 6 | 750 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2RDY_A | 1.12e-19 | 289 | 725 | 327 | 789 | ChainA, BH0842 protein [Halalkalibacterium halodurans C-125],2RDY_B Chain B, BH0842 protein [Halalkalibacterium halodurans C-125] |
| 7KMQ_A | 9.67e-13 | 300 | 683 | 359 | 752 | ChainA, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306],7KMQ_B Chain B, Glyco_hyd_65N_2 domain-containing protein [Xanthomonas citri pv. citri str. 306] |
| 4UFC_A | 9.85e-13 | 242 | 683 | 257 | 736 | Crystalstructure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus],4UFC_B Crystal structure of the GH95 enzyme BACOVA_03438 [Bacteroides ovatus] |
| 2EAB_A | 1.90e-10 | 252 | 688 | 350 | 849 | Crystalstructure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAB_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum (apo form) [Bifidobacterium bifidum],2EAC_A Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum],2EAC_B Crystal structure of 1,2-a-L-fucosidase from Bifidobacterium bifidum in complex with deoxyfuconojirimycin [Bifidobacterium bifidum] |
| 2EAE_A | 4.31e-10 | 252 | 688 | 349 | 848 | ChainA, Alpha-fucosidase [Bifidobacterium bifidum] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
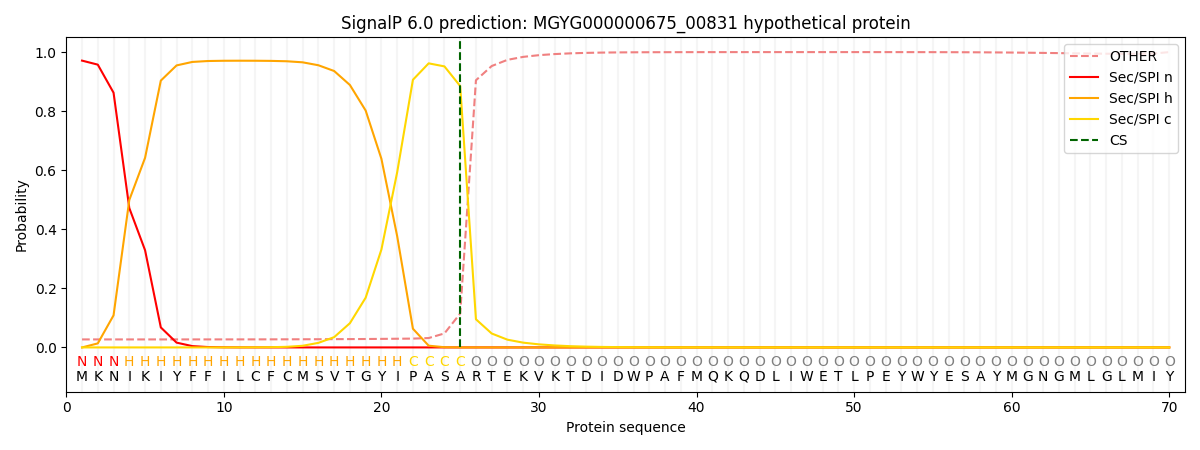
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.045111 | 0.949415 | 0.003159 | 0.000823 | 0.000624 | 0.000857 |