You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000675_00832
You are here: Home > Sequence: MGYG000000675_00832
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides congonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides congonensis | |||||||||||
| CAZyme ID | MGYG000000675_00832 | |||||||||||
| CAZy Family | GH88 | |||||||||||
| CAZyme Description | Unsaturated chondroitin disaccharide hydrolase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 74047; End: 75270 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH88 | 73 | 398 | 7.3e-121 | 0.9756838905775076 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam07470 | Glyco_hydro_88 | 4.67e-09 | 74 | 303 | 28 | 234 | Glycosyl Hydrolase Family 88. Unsaturated glucuronyl hydrolase catalyzes the hydrolytic release of unsaturated glucuronic acids from oligosaccharides (EC:3.2.1.-) produced by the reactions of polysaccharide lyases. |
| pfam16386 | DUF4995 | 0.002 | 1 | 54 | 1 | 53 | Domain of unknown function. This family around 100 residues locates in the N-terminal of some uncharacterized proteins and glucuronyl hydrolases in various Bacteroides species. The function of this family remains unknown. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUU06876.1 | 9.48e-303 | 1 | 407 | 1 | 407 |
| QQT78393.1 | 3.86e-302 | 1 | 407 | 1 | 407 |
| QRP56362.1 | 3.86e-302 | 1 | 407 | 1 | 407 |
| ASM67776.1 | 1.84e-292 | 15 | 407 | 1 | 393 |
| QKH85475.1 | 4.47e-229 | 1 | 404 | 1 | 403 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3WIW_A | 5.27e-101 | 1 | 404 | 1 | 393 | Crystalstructure of unsaturated glucuronyl hydrolase specific for heparin [Pedobacter heparinus DSM 2366] |
| 2ZZR_A | 2.59e-49 | 31 | 398 | 28 | 388 | Crystalstructure of unsaturated glucuronyl hydrolase from Streptcoccus agalactiae [Streptococcus agalactiae] |
| 3ANJ_A | 2.65e-49 | 31 | 398 | 29 | 389 | Crystalstructure of unsaturated glucuronyl hydrolase from Streptcoccus agalactiae [Streptococcus agalactiae serogroup III] |
| 3WUX_A | 3.71e-49 | 31 | 398 | 29 | 389 | Crystalstructure of unsaturated glucuronyl hydrolase mutant D115N/K370S from Streptococcus agalactiae [Streptococcus agalactiae NEM316] |
| 3ANI_A | 1.41e-48 | 31 | 398 | 29 | 389 | Crystalstructure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae [Streptococcus agalactiae serogroup III],3ANK_A Crystal structure of unsaturated glucuronyl hydrolase mutant D175N from Streptcoccus agalactiae complexed with dGlcA-GalNAc6S [Streptococcus agalactiae serogroup III] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KLZ3 | 3.20e-107 | 74 | 402 | 76 | 397 | Unsaturated glucuronyl hydrolase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_21900 PE=1 SV=1 |
| Q8E372 | 1.45e-48 | 31 | 398 | 29 | 389 | Unsaturated chondroitin disaccharide hydrolase OS=Streptococcus agalactiae serotype III (strain NEM316) OX=211110 GN=gbs1889 PE=1 SV=1 |
| Q9A0T3 | 3.13e-46 | 31 | 398 | 30 | 390 | Unsaturated chondroitin disaccharide hydrolase OS=Streptococcus pyogenes serotype M1 OX=301447 GN=ugl PE=1 SV=1 |
| Q8DR77 | 1.23e-41 | 31 | 398 | 27 | 387 | Unsaturated chondroitin disaccharide hydrolase OS=Streptococcus pneumoniae (strain ATCC BAA-255 / R6) OX=171101 GN=ugl PE=1 SV=1 |
| Q9RC92 | 8.58e-40 | 73 | 398 | 40 | 363 | Unsaturated glucuronyl hydrolase OS=Bacillus sp. (strain GL1) OX=84635 GN=ugl PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
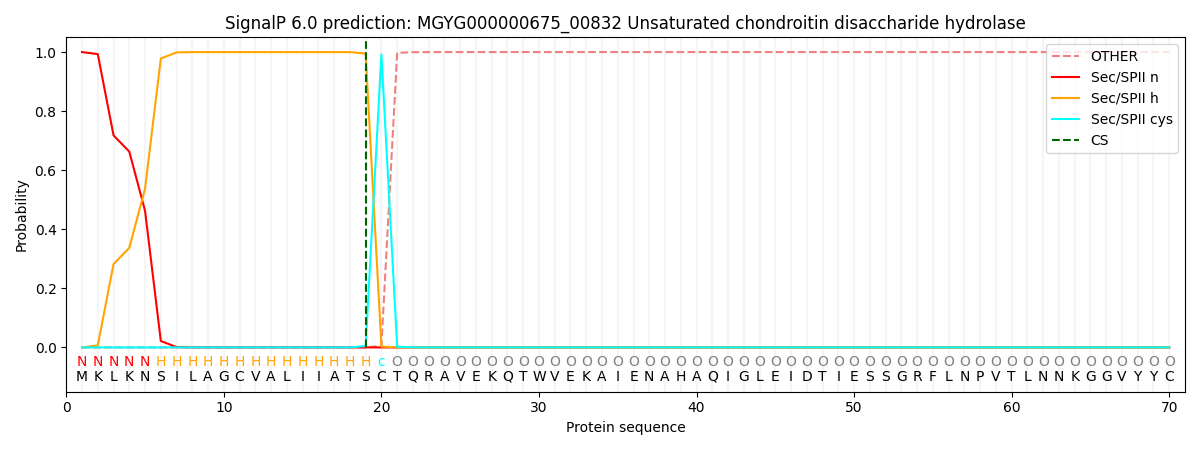
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000001 | 1.000058 | 0.000000 | 0.000000 | 0.000000 |
