You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000675_01615
You are here: Home > Sequence: MGYG000000675_01615
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides congonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides congonensis | |||||||||||
| CAZyme ID | MGYG000000675_01615 | |||||||||||
| CAZy Family | GH89 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 5275; End: 9102 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH123 | 740 | 1274 | 5.2e-208 | 0.9907063197026023 |
| GH89 | 58 | 698 | 7.6e-203 | 0.9803921568627451 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05089 | NAGLU | 3.21e-148 | 117 | 438 | 1 | 333 | Alpha-N-acetylglucosaminidase (NAGLU) tim-barrel domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This central domain has a tim barrel fold. |
| pfam12972 | NAGLU_C | 7.27e-64 | 446 | 689 | 1 | 242 | Alpha-N-acetylglucosaminidase (NAGLU) C-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This C-terminal domain has an all alpha helical fold. |
| pfam13320 | DUF4091 | 8.82e-25 | 1171 | 1231 | 1 | 66 | Domain of unknown function (DUF4091). This presumed domain is functionally uncharacterized. This domain family is found in bacteria, archaea and eukaryotes, and is approximately 70 amino acids in length. There is a single completely conserved residue G that may be functionally important. |
| pfam12971 | NAGLU_N | 1.10e-23 | 24 | 102 | 1 | 81 | Alpha-N-acetylglucosaminidase (NAGLU) N-terminal domain. Alpha-N-acetylglucosaminidase, a lysosomal enzyme required for the stepwise degradation of heparan sulfate. Mutations on the alpha-N-acetylglucosaminidase (NAGLU) gene can lead to Mucopolysaccharidosis type IIIB (MPS IIIB; or Sanfilippo syndrome type B) characterized by neurological dysfunction but relatively mild somatic manifestations. The structure shows that the enzyme is composed of three domains. This N-terminal domain has an alpha-beta fold. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQT78717.1 | 0.0 | 3 | 709 | 5 | 713 |
| QRP59399.1 | 0.0 | 3 | 709 | 5 | 713 |
| ASM64780.1 | 0.0 | 3 | 709 | 5 | 713 |
| QUU06381.1 | 0.0 | 3 | 709 | 5 | 713 |
| QIU92982.1 | 0.0 | 20 | 709 | 23 | 713 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5L7V_A | 1.56e-190 | 743 | 1275 | 29 | 563 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7V_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 5L7R_A | 2.60e-190 | 743 | 1275 | 44 | 578 | ChainA, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7R_B Chain B, glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_A Chain A, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482],5L7U_B Chain B, Glycoside hydrolase [Phocaeicola vulgatus ATCC 8482] |
| 2VC9_A | 3.31e-127 | 57 | 688 | 209 | 849 | Family89 Glycoside Hydrolase from Clostridium perfringens in complex with 2-acetamido-1,2-dideoxynojirmycin [Clostridium perfringens],2VCA_A Family 89 glycoside hydrolase from Clostridium perfringens in complex with beta-N-acetyl-D-glucosamine [Clostridium perfringens],2VCB_A Family 89 Glycoside Hydrolase from Clostridium perfringens in complex with PUGNAc [Clostridium perfringens],2VCC_A Family 89 Glycoside Hydrolase from Clostridium perfringens [Clostridium perfringens] |
| 7MFK_A | 4.02e-127 | 57 | 688 | 217 | 857 | ChainA, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124],7MFL_A Chain A, Alpha-N-acetylglucosaminidase family protein [Clostridium perfringens ATCC 13124] |
| 4A4A_A | 4.15e-126 | 57 | 688 | 232 | 872 | CpGH89(E483Q, E601Q), from Clostridium perfringens, in complex with its substrate GlcNAc-alpha-1,4-galactose [Clostridium perfringens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q9FNA3 | 7.75e-89 | 24 | 686 | 47 | 771 | Alpha-N-acetylglucosaminidase OS=Arabidopsis thaliana OX=3702 GN=NAGLU PE=2 SV=1 |
| P54802 | 8.36e-82 | 20 | 664 | 26 | 682 | Alpha-N-acetylglucosaminidase OS=Homo sapiens OX=9606 GN=NAGLU PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
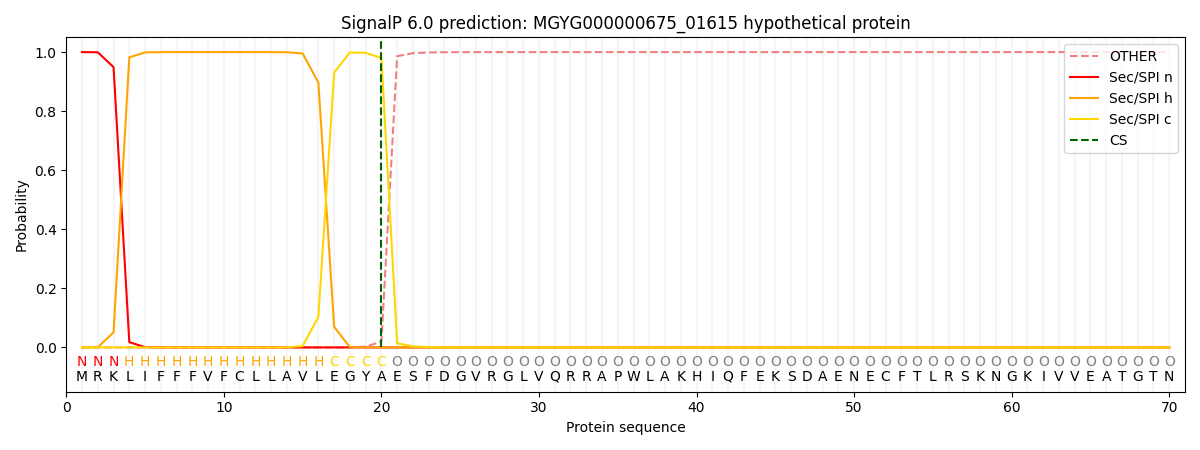
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000183 | 0.999229 | 0.000148 | 0.000144 | 0.000131 | 0.000129 |
