You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000675_03090
You are here: Home > Sequence: MGYG000000675_03090
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Bacteroides congonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides congonensis | |||||||||||
| CAZyme ID | MGYG000000675_03090 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11692; End: 14040 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 60 | 426 | 3.4e-82 | 0.990228013029316 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 1.17e-50 | 61 | 426 | 4 | 316 | Glycosyl hydrolases family 35. |
| PLN03059 | PLN03059 | 4.27e-24 | 51 | 425 | 29 | 339 | beta-galactosidase; Provisional |
| COG1874 | GanA | 2.73e-17 | 52 | 205 | 1 | 160 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 7.60e-09 | 77 | 205 | 6 | 137 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| smart01029 | BetaGal_dom2 | 3.25e-04 | 490 | 614 | 52 | 171 | Beta-galactosidase, domain 2. This is the second domain of the five-domain beta-galactosidase enzyme that altogether catalyses the hydrolysis of beta(1-3) and beta(1-4) galactosyl bonds in oligosaccharides as well as the inverse reaction of enzymatic condensation and trans-glycosylation. This domain is made up of 16 antiparallel beta-strands and an alpha-helix at its C terminus. The fold of this domain appears to be unique. In addition, the last seven strands of the domain form a subdomain with an immunoglobulin-like (I-type Ig) fold in which the first strand is divided between the two beta-sheets. In penicillin spp this strand is interrupted by a 12-residue insertion which forms an additional edge-strand to the second beta-sheet of the sub-domain. The remainder of the second domain forms a series of beta-hairpins at its N terminus, four strands of which are contiguous with part of the Ig-like sub-domain, forming in total a seven-stranded antiparallel beta-sheet. This domain is associated with family Glyco_hydro_35, which is N-terminal to it, but itself has no metazoan members. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QDM09602.1 | 0.0 | 1 | 781 | 1 | 781 |
| QUT80076.1 | 0.0 | 1 | 781 | 1 | 781 |
| QGT71913.1 | 0.0 | 1 | 782 | 1 | 782 |
| QIU94789.1 | 0.0 | 1 | 780 | 1 | 780 |
| QVJ80561.1 | 0.0 | 1 | 780 | 1 | 783 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6EON_A | 5.42e-28 | 45 | 204 | 21 | 181 | GalactanaseBT0290 [Bacteroides thetaiotaomicron VPI-5482] |
| 3D3A_A | 1.60e-27 | 48 | 205 | 4 | 162 | Crystalstructure of a beta-galactosidase from Bacteroides thetaiotaomicron [Bacteroides thetaiotaomicron VPI-5482] |
| 7KDV_A | 3.00e-26 | 62 | 205 | 28 | 172 | ChainA, Beta-galactosidase [Mus musculus],7KDV_C Chain C, Beta-galactosidase [Mus musculus],7KDV_E Chain E, Beta-galactosidase [Mus musculus],7KDV_G Chain G, Beta-galactosidase [Mus musculus],7KDV_I Chain I, Beta-galactosidase [Mus musculus],7KDV_K Chain K, Beta-galactosidase [Mus musculus] |
| 4E8C_A | 7.85e-26 | 61 | 444 | 12 | 342 | Crystalstructure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8C_B Crystal structure of streptococcal beta-galactosidase in complex with galactose [Streptococcus pneumoniae TIGR4],4E8D_A Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4],4E8D_B Crystal structure of streptococcal beta-galactosidase [Streptococcus pneumoniae TIGR4] |
| 3THC_A | 7.37e-25 | 62 | 205 | 21 | 165 | Crystalstructure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_B Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_C Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THC_D Crystal structure of human beta-galactosidase in complex with galactose [Homo sapiens],3THD_A Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_B Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_C Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens],3THD_D Crystal structure of human beta-galactosidase in complex with 1-deoxygalactonojirimycin [Homo sapiens] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q8GX69 | 5.99e-28 | 51 | 426 | 24 | 336 | Beta-galactosidase 16 OS=Arabidopsis thaliana OX=3702 GN=BGAL16 PE=2 SV=2 |
| Q58D55 | 3.46e-27 | 62 | 205 | 43 | 187 | Beta-galactosidase OS=Bos taurus OX=9913 GN=GLB1 PE=2 SV=1 |
| P48982 | 8.09e-27 | 62 | 205 | 40 | 184 | Beta-galactosidase OS=Xanthomonas manihotis OX=43353 GN=bga PE=1 SV=1 |
| Q9TRY9 | 3.53e-26 | 62 | 205 | 45 | 189 | Beta-galactosidase OS=Canis lupus familiaris OX=9615 GN=GLB1 PE=1 SV=3 |
| Q93Z24 | 6.87e-26 | 62 | 424 | 73 | 392 | Beta-galactosidase 17 OS=Arabidopsis thaliana OX=3702 GN=BGAL17 PE=2 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
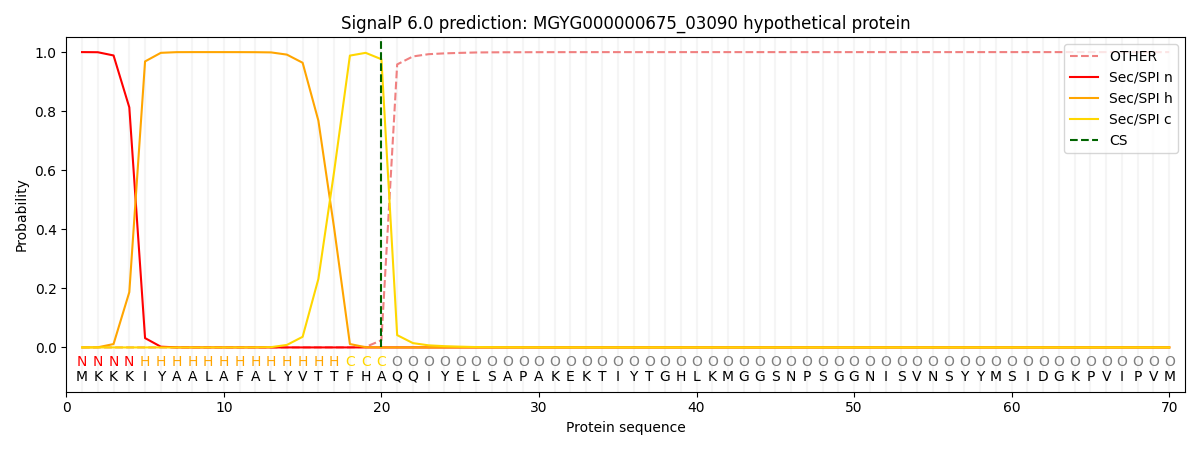
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000487 | 0.998596 | 0.000306 | 0.000198 | 0.000206 | 0.000178 |
