You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000675_03522
You are here: Home > Sequence: MGYG000000675_03522
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
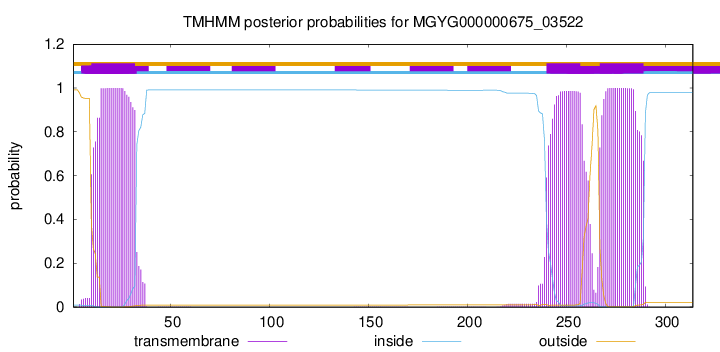
TMHMM annotations
Basic Information help
| Species | Bacteroides congonensis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Bacteroides; Bacteroides congonensis | |||||||||||
| CAZyme ID | MGYG000000675_03522 | |||||||||||
| CAZy Family | GT0 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2826; End: 3770 Strand: - | |||||||||||
Full Sequence Download help
| MDSYIYILDD LVFFFTGVLF LYLFILAVAS HFKHIIYSKT KKKYHCAILI PEGSPLITIY | 60 |
| KEEPYEFITY NDLYQRINSL DKEQYDLVLI LSDTACALSP RLLDKIYDAY DSGIQAIQLH | 120 |
| SITENRQGFR NRFRILCDEI KNSICRAGNT QFGLSSNLLG TNMAIDLEWL QKNLKSSKTN | 180 |
| IERKLLRQNI YIDYLPDAIV YCQSSPVCPY RKRIRKMASY LLPSLLEGNW SFFNRIIQQL | 240 |
| IPSPLKLCIF VGIWALLITG YDWTSSFSWW ILLFGLLITY SLAIPDYLVE DKKKKKHSIW | 300 |
| RKRHLNSELK EIPA | 314 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13632 | Glyco_trans_2_3 | 7.83e-11 | 87 | 282 | 1 | 194 | Glycosyl transferase family group 2. Members of this family of prokaryotic proteins include putative glucosyltransferases, which are involved in bacterial capsule biosynthesis. |
| pfam13641 | Glyco_tranf_2_3 | 0.004 | 85 | 204 | 88 | 213 | Glycosyltransferase like family 2. Members of this family of prokaryotic proteins include putative glucosyltransferase, which are involved in bacterial capsule biosynthesis. |
| cd06438 | EpsO_like | 0.008 | 82 | 173 | 79 | 170 | EpsO protein participates in the methanolan synthesis. The Methylobacillus sp EpsO protein is predicted to participate in the methanolan synthesis. Methanolan is an exopolysaccharide (EPS), composed of glucose, mannose and galactose. A 21 genes cluster was predicted to participate in the methanolan synthesis. Gene disruption analysis revealed that EpsO is one of the glycosyltransferase enzymes involved in the synthesis of repeating sugar units onto the lipid carrier. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIU96407.1 | 1.08e-220 | 1 | 314 | 1 | 314 |
| QUT24338.1 | 1.25e-185 | 1 | 314 | 2 | 315 |
| QDH56310.1 | 1.46e-184 | 1 | 314 | 2 | 315 |
| QDM07248.1 | 1.46e-184 | 1 | 314 | 2 | 315 |
| QUT81293.1 | 1.46e-184 | 1 | 314 | 2 | 315 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
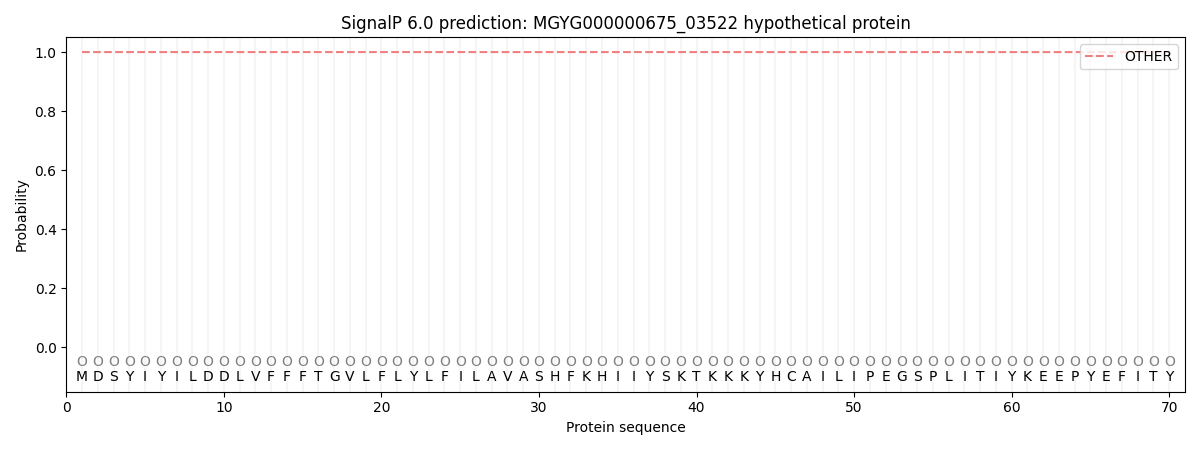
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000067 | 0.000003 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |