You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000681_00382
You are here: Home > Sequence: MGYG000000681_00382
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Lactococcus_A raffinolactis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes; Bacilli; Lactobacillales; Streptococcaceae; Lactococcus_A; Lactococcus_A raffinolactis | |||||||||||
| CAZyme ID | MGYG000000681_00382 | |||||||||||
| CAZy Family | GH73 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 1208; End: 2746 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam18013 | Phage_lysozyme2 | 2.45e-33 | 241 | 378 | 1 | 139 | Phage tail lysozyme. This domain has a lysozyme like fold. It is found in the tail protein of various phages probably giving them the ability to degrade the host cell wall peptidoglycan layer. |
| PRK06347 | PRK06347 | 1.30e-21 | 33 | 212 | 400 | 592 | 1,4-beta-N-acetylmuramoylhydrolase. |
| PRK06347 | PRK06347 | 2.67e-21 | 14 | 216 | 306 | 528 | 1,4-beta-N-acetylmuramoylhydrolase. |
| COG3942 | COG3942 | 2.04e-20 | 395 | 509 | 58 | 171 | Surface antigen [Cell wall/membrane/envelope biogenesis]. |
| pfam05257 | CHAP | 7.16e-17 | 401 | 487 | 1 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIW61582.1 | 0.0 | 1 | 512 | 1 | 512 |
| QIW52867.1 | 0.0 | 1 | 512 | 1 | 512 |
| ATC61235.1 | 0.0 | 1 | 512 | 1 | 512 |
| QIW56964.1 | 0.0 | 1 | 512 | 1 | 512 |
| QIW52488.1 | 0.0 | 1 | 512 | 1 | 512 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2K3A_A | 1.75e-15 | 400 | 511 | 47 | 155 | ChainA, CHAP domain protein [Staphylococcus saprophyticus subsp. saprophyticus ATCC 15305 = NCTC 7292] |
| 2LRJ_A | 5.33e-15 | 401 | 510 | 7 | 113 | ChainA, Staphyloxanthin biosynthesis protein, putative [Staphylococcus aureus subsp. aureus COL] |
| 4B8V_A | 1.24e-10 | 41 | 211 | 44 | 216 | ChainA, Extracellular Protein 6 [Fulvia fulva],4B9H_A Chain A, Extracellular Protein 6 [Fulvia fulva] |
| 5K2L_A | 1.25e-06 | 40 | 81 | 4 | 45 | Crystalstructure of LysM domain from Volvox carteri chitinase [Volvox carteri f. nagariensis],5YZK_A Solution structure of LysM domain from a chitinase derived from Volvox carteri [Volvox carteri f. nagariensis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P39046 | 2.66e-15 | 40 | 212 | 488 | 665 | Muramidase-2 OS=Enterococcus hirae (strain ATCC 9790 / DSM 20160 / JCM 8729 / LMG 6399 / NBRC 3181 / NCIMB 6459 / NCDO 1258 / NCTC 12367 / WDCM 00089 / R) OX=768486 GN=EHR_05900 PE=1 SV=1 |
| O07532 | 4.16e-14 | 40 | 211 | 175 | 350 | Peptidoglycan endopeptidase LytF OS=Bacillus subtilis (strain 168) OX=224308 GN=lytF PE=1 SV=2 |
| O31852 | 2.28e-13 | 14 | 220 | 2 | 209 | D-gamma-glutamyl-meso-diaminopimelic acid endopeptidase CwlS OS=Bacillus subtilis (strain 168) OX=224308 GN=cwlS PE=1 SV=1 |
| Q7CCJ3 | 7.70e-13 | 392 | 509 | 143 | 257 | Staphylococcal secretory antigen SsaA OS=Staphylococcus epidermidis (strain ATCC 12228 / FDA PCI 1200) OX=176280 GN=ssaA PE=3 SV=1 |
| Q5HLV2 | 7.70e-13 | 392 | 509 | 143 | 257 | Staphylococcal secretory antigen SsaA OS=Staphylococcus epidermidis (strain ATCC 35984 / RP62A) OX=176279 GN=ssaA1 PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
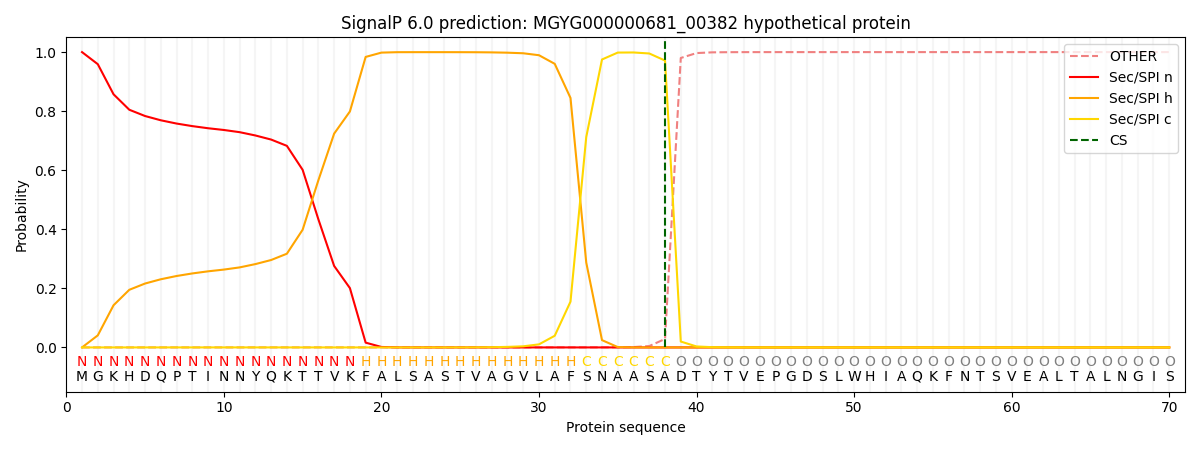
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000355 | 0.998970 | 0.000169 | 0.000191 | 0.000162 | 0.000140 |
