You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000682_00875
You are here: Home > Sequence: MGYG000000682_00875
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA4334; | |||||||||||
| CAZyme ID | MGYG000000682_00875 | |||||||||||
| CAZy Family | GH35 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 8404; End: 8829 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH35 | 30 | 139 | 3.7e-45 | 0.3550488599348534 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam01301 | Glyco_hydro_35 | 6.12e-34 | 29 | 141 | 1 | 114 | Glycosyl hydrolases family 35. |
| PLN03059 | PLN03059 | 2.42e-24 | 3 | 139 | 10 | 147 | beta-galactosidase; Provisional |
| COG1874 | GanA | 2.89e-12 | 26 | 131 | 4 | 101 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam02449 | Glyco_hydro_42 | 6.06e-05 | 50 | 131 | 8 | 79 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| ADY37835.1 | 1.17e-61 | 22 | 141 | 27 | 146 |
| AGB28883.1 | 1.05e-55 | 20 | 139 | 342 | 461 |
| QVJ80561.1 | 8.39e-44 | 34 | 139 | 62 | 167 |
| ADE81979.1 | 8.39e-44 | 34 | 139 | 62 | 167 |
| BCA48705.1 | 7.11e-43 | 26 | 131 | 40 | 145 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6IK6_A | 1.21e-24 | 22 | 139 | 7 | 125 | ChainA, Beta-galactosidase [Solanum lycopersicum],6IK6_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK7_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK7_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK8_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK8_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
| 3W5F_A | 1.21e-24 | 22 | 139 | 7 | 125 | ChainA, Beta-galactosidase [Solanum lycopersicum],3W5F_B Chain B, Beta-galactosidase [Solanum lycopersicum],3W5G_A Chain A, Beta-galactosidase [Solanum lycopersicum],3W5G_B Chain B, Beta-galactosidase [Solanum lycopersicum],6IK5_A Chain A, Beta-galactosidase [Solanum lycopersicum],6IK5_B Chain B, Beta-galactosidase [Solanum lycopersicum] |
| 5GSL_A | 1.07e-23 | 22 | 128 | 5 | 115 | Glycosidehydrolase A [Pyrococcus horikoshii OT3],5GSL_B Glycoside hydrolase A [Pyrococcus horikoshii OT3] |
| 5GSM_A | 3.70e-23 | 22 | 128 | 3 | 113 | Glycosidehydrolase B with product [Thermococcus kodakarensis KOD1],5GSM_B Glycoside hydrolase B with product [Thermococcus kodakarensis KOD1] |
| 6EON_A | 2.48e-20 | 29 | 139 | 34 | 145 | GalactanaseBT0290 [Bacteroides thetaiotaomicron VPI-5482] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q6ZJJ0 | 3.48e-26 | 23 | 128 | 33 | 138 | Beta-galactosidase 11 OS=Oryza sativa subsp. japonica OX=39947 GN=Os08g0549200 PE=2 SV=1 |
| Q5Z7L0 | 1.23e-23 | 16 | 139 | 15 | 139 | Beta-galactosidase 9 OS=Oryza sativa subsp. japonica OX=39947 GN=Os06g0573600 PE=2 SV=1 |
| Q9SCV1 | 1.71e-23 | 21 | 128 | 39 | 146 | Beta-galactosidase 11 OS=Arabidopsis thaliana OX=3702 GN=BGAL11 PE=2 SV=1 |
| Q9FFN4 | 2.29e-23 | 21 | 128 | 30 | 137 | Beta-galactosidase 6 OS=Arabidopsis thaliana OX=3702 GN=BGAL6 PE=2 SV=1 |
| Q8GX69 | 3.17e-23 | 22 | 139 | 24 | 142 | Beta-galactosidase 16 OS=Arabidopsis thaliana OX=3702 GN=BGAL16 PE=2 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
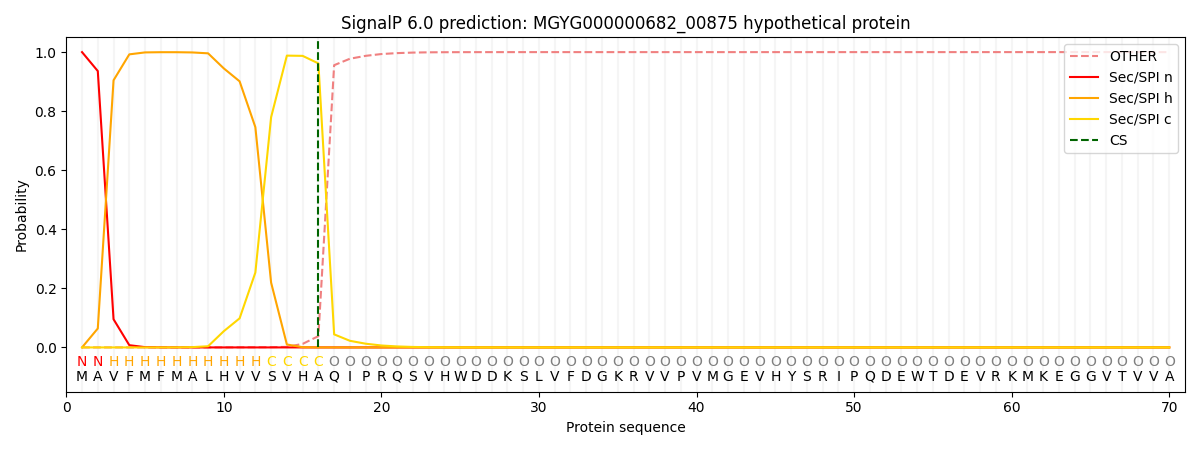
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000725 | 0.998383 | 0.000190 | 0.000249 | 0.000216 | 0.000196 |
