You are browsing environment: HUMAN GUT
MGYG000000682_01619
Basic Information
help
Species
Lineage
Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA4334;
CAZyme ID
MGYG000000682_01619
CAZy Family
GH128
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
960
106577.45
5.2347
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000682
2426258
MAG
Kazakhstan
Asia
Gene Location
Start: 20277;
End: 23159
Strand: +
No EC number prediction in MGYG000000682_01619.
Family
Start
End
Evalue
family coverage
GH128
287
442
9.4e-17
0.7008928571428571
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
pfam11790
Glyco_hydro_cc
1.12e-06
277
442
71
235
Glycosyl hydrolase catalytic core. This family is probably a glycosyl hydrolase, and is conserved in fungi and some Proteobacteria. The pombe member is annotated as being from IPR013781.
more
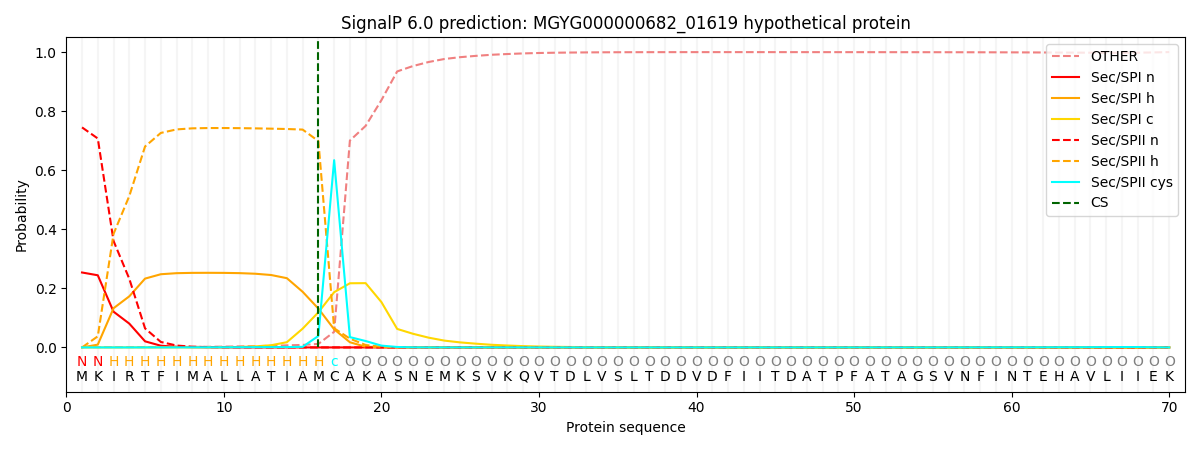
This protein is predicted as LIPO
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.002124
0.246415
0.750780
0.000276
0.000201
0.000186
There is no transmembrane helices in MGYG000000682_01619.