You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000683_00580
You are here: Home > Sequence: MGYG000000683_00580
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-977 sp000434295 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Proteobacteria; Alphaproteobacteria; RF32; CAG-977; CAG-977; CAG-977 sp000434295 | |||||||||||
| CAZyme ID | MGYG000000683_00580 | |||||||||||
| CAZy Family | GT2 | |||||||||||
| CAZyme Description | Glucans biosynthesis glucosyltransferase H | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 39053; End: 41098 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG2943 | MdoH | 0.0 | 14 | 639 | 63 | 692 | Membrane glycosyltransferase [Cell wall/membrane/envelope biogenesis, Carbohydrate transport and metabolism]. |
| PRK05454 | PRK05454 | 0.0 | 10 | 570 | 36 | 599 | glucans biosynthesis glucosyltransferase MdoH. |
| cd04191 | Glucan_BSP_MdoH | 4.80e-150 | 97 | 350 | 1 | 254 | Glucan_BSP_MdoH catalyzes the elongation of beta-1,2 polyglucose chains of glucan. Periplasmic Glucan Biosynthesis protein MdoH is a glucosyltransferase that catalyzes the elongation of beta-1,2 polyglucose chains of glucan, requiring a beta-glucoside as a primer and UDP-glucose as a substrate. Glucans are composed of 5 to 10 units of glucose forming a highly branched structure, where beta-1,2-linked glucose constitutes a linear backbone to which branches are attached by beta-1,6 linkages. In Escherichia coli, glucans are located in the periplasmic space, functioning as regulator of osmolarity. It is synthesized at a maximum when cells are grown in a medium with low osmolarity. It has been shown to span the cytoplasmic membrane. |
| cd06421 | CESA_CelA_like | 6.95e-17 | 102 | 351 | 8 | 234 | CESA_CelA_like are involved in the elongation of the glucan chain of cellulose. Family of proteins related to Agrobacterium tumefaciens CelA and Gluconacetobacter xylinus BscA. These proteins are involved in the elongation of the glucan chain of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues. They are putative catalytic subunit of cellulose synthase, which is a glycosyltransferase using UDP-glucose as the substrate. The catalytic subunit is an integral membrane protein with 6 transmembrane segments and it is postulated that the protein is anchored in the membrane at the N-terminal end. |
| cd06423 | CESA_like | 8.31e-15 | 99 | 281 | 1 | 168 | CESA_like is the cellulose synthase superfamily. The cellulose synthase (CESA) superfamily includes a wide variety of glycosyltransferase family 2 enzymes that share the common characteristic of catalyzing the elongation of polysaccharide chains. The members include cellulose synthase catalytic subunit, chitin synthase, glucan biosynthesis protein and other families of CESA-like proteins. Cellulose synthase catalyzes the polymerization reaction of cellulose, an aggregate of unbranched polymers of beta-1,4-linked glucose residues in plants, most algae, some bacteria and fungi, and even some animals. In bacteria, algae and lower eukaryotes, there is a second unrelated type of cellulose synthase (Type II), which produces acylated cellulose, a derivative of cellulose. Chitin synthase catalyzes the incorporation of GlcNAc from substrate UDP-GlcNAc into chitin, which is a linear homopolymer of beta-(1,4)-linked GlcNAc residues and Glucan Biosynthesis protein catalyzes the elongation of beta-1,2 polyglucose chains of Glucan. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QRO00644.1 | 6.58e-199 | 14 | 674 | 13 | 670 |
| ATB33264.1 | 2.44e-191 | 25 | 674 | 24 | 670 |
| QSQ18747.1 | 6.91e-191 | 16 | 671 | 15 | 664 |
| QPM81905.1 | 7.87e-191 | 14 | 671 | 13 | 668 |
| QDE96388.1 | 7.87e-191 | 14 | 671 | 13 | 668 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q0HJ63 | 7.80e-165 | 16 | 669 | 57 | 710 | Glucans biosynthesis glucosyltransferase H OS=Shewanella sp. (strain MR-4) OX=60480 GN=opgH PE=3 SV=1 |
| Q0HUS0 | 2.20e-164 | 16 | 669 | 57 | 710 | Glucans biosynthesis glucosyltransferase H OS=Shewanella sp. (strain MR-7) OX=60481 GN=opgH PE=3 SV=1 |
| A0KWF0 | 4.38e-164 | 16 | 669 | 57 | 710 | Glucans biosynthesis glucosyltransferase H OS=Shewanella sp. (strain ANA-3) OX=94122 GN=opgH PE=3 SV=1 |
| Q9KSG9 | 1.15e-162 | 16 | 641 | 56 | 679 | Glucans biosynthesis glucosyltransferase H OS=Vibrio cholerae serotype O1 (strain ATCC 39315 / El Tor Inaba N16961) OX=243277 GN=opgH PE=3 SV=1 |
| A5F1Q0 | 1.15e-162 | 16 | 641 | 56 | 679 | Glucans biosynthesis glucosyltransferase H OS=Vibrio cholerae serotype O1 (strain ATCC 39541 / Classical Ogawa 395 / O395) OX=345073 GN=opgH PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER

| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000031 | 0.000000 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |
TMHMM Annotations download full data without filtering help
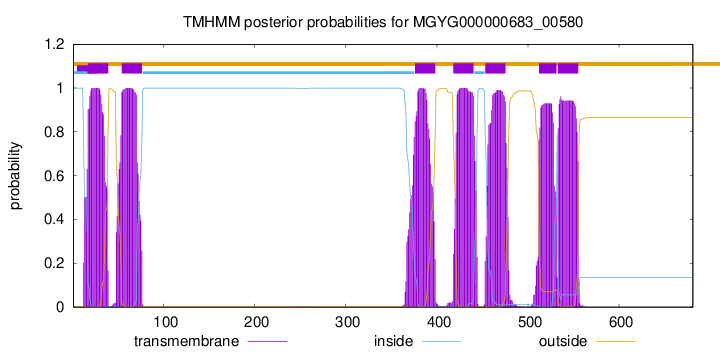
| start | end |
|---|---|
| 17 | 39 |
| 54 | 76 |
| 376 | 398 |
| 418 | 440 |
| 453 | 475 |
| 512 | 531 |
| 533 | 555 |
