You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000685_00483
You are here: Home > Sequence: MGYG000000685_00483
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | CAG-590 sp900552885 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Lachnospirales; Lachnospiraceae; CAG-590; CAG-590 sp900552885 | |||||||||||
| CAZyme ID | MGYG000000685_00483 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28315; End: 29652 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 50 | 299 | 2.9e-92 | 0.9873417721518988 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 1.68e-70 | 50 | 306 | 4 | 271 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 2.96e-09 | 65 | 245 | 69 | 227 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
| cd01304 | FMDH_A | 0.001 | 183 | 233 | 273 | 322 | Formylmethanofuran dehydrogenase (FMDH) subunit A; Methanogenic bacteria and archea derive the energy for autotrophic growth from methanogenesis, the reduction of CO2 with molecular hydrogen as the electron donor. FMDH catalyzes the first step in methanogenesis, the formyl-methanofuran synthesis. In this step, CO2 is bound to methanofuran and subsequently reduced to the formyl state with electrons derived from hydrogen. |
| COG1229 | FwdA | 0.003 | 85 | 233 | 181 | 333 | Formylmethanofuran dehydrogenase subunit A [Energy production and conversion]. |
| cd00063 | FN3 | 0.004 | 381 | 427 | 32 | 80 | Fibronectin type 3 domain; One of three types of internal repeats found in the plasma protein fibronectin. Its tenth fibronectin type III repeat contains an RGD cell recognition sequence in a flexible loop between 2 strands. Approximately 2% of all animal proteins contain the FN3 repeat; including extracellular and intracellular proteins, membrane spanning cytokine receptors, growth hormone receptors, tyrosine phosphatase receptors, and adhesion molecules. FN3-like domains are also found in bacterial glycosyl hydrolases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QWT53734.1 | 6.05e-118 | 32 | 338 | 215 | 511 |
| QNM00780.1 | 2.42e-117 | 32 | 338 | 215 | 511 |
| CBK83877.1 | 1.05e-107 | 35 | 335 | 231 | 523 |
| QBR94633.1 | 1.77e-107 | 25 | 336 | 81 | 383 |
| AFA47670.1 | 5.72e-106 | 32 | 336 | 199 | 495 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6GJF_A | 9.43e-103 | 32 | 338 | 7 | 299 | Ancestralendocellulase Cel5A [synthetic construct],6GJF_B Ancestral endocellulase Cel5A [synthetic construct],6GJF_C Ancestral endocellulase Cel5A [synthetic construct],6GJF_D Ancestral endocellulase Cel5A [synthetic construct],6GJF_E Ancestral endocellulase Cel5A [synthetic construct],6GJF_F Ancestral endocellulase Cel5A [synthetic construct] |
| 3PZT_A | 7.02e-95 | 27 | 335 | 22 | 320 | Structureof the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZT_B Structure of the endo-1,4-beta-glucanase from Bacillus subtilis 168 with manganese(II) ion [Bacillus subtilis subsp. subtilis str. 168],3PZU_A P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZU_B P212121 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_A C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_B C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_C C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168],3PZV_D C2 crystal form of the endo-1,4-beta-glucanase from Bacillus subtilis 168 [Bacillus subtilis subsp. subtilis str. 168] |
| 4XZW_A | 4.54e-92 | 32 | 335 | 6 | 298 | Endo-glucanasechimera C10 [uncultured bacterium] |
| 4XZB_A | 3.78e-88 | 32 | 335 | 6 | 299 | endo-glucanaseGsCelA P1 [Geobacillus sp. 70PC53] |
| 1LF1_A | 5.57e-86 | 32 | 335 | 6 | 296 | CrystalStructure of Cel5 from Alkalophilic Bacillus sp. [Bacillus subtilis] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P22541 | 5.80e-97 | 23 | 338 | 101 | 406 | Endoglucanase A OS=Butyrivibrio fibrisolvens OX=831 GN=celA PE=1 SV=1 |
| P15704 | 1.28e-94 | 26 | 335 | 35 | 331 | Endoglucanase OS=Clostridium saccharobutylicum OX=169679 GN=eglA PE=3 SV=1 |
| P07983 | 3.94e-94 | 22 | 393 | 22 | 404 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=2 |
| P10475 | 2.20e-93 | 22 | 393 | 22 | 404 | Endoglucanase OS=Bacillus subtilis (strain 168) OX=224308 GN=eglS PE=1 SV=1 |
| P23549 | 7.56e-91 | 22 | 335 | 22 | 325 | Endoglucanase OS=Bacillus subtilis OX=1423 GN=bglC PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
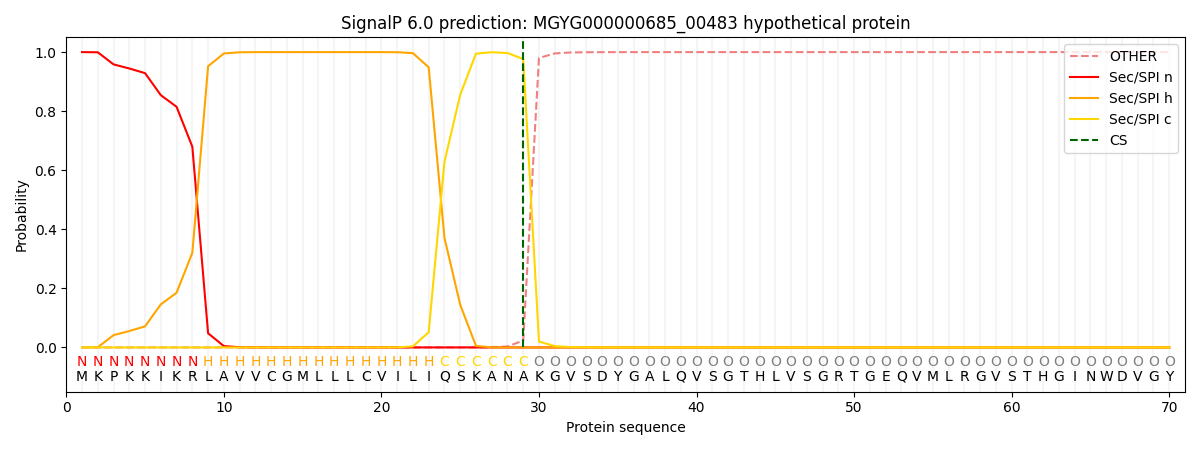
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000289 | 0.998857 | 0.000314 | 0.000185 | 0.000170 | 0.000163 |
