You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000692_00566
You are here: Home > Sequence: MGYG000000692_00566
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotellamassilia sp900543155 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotellamassilia; Prevotellamassilia sp900543155 | |||||||||||
| CAZyme ID | MGYG000000692_00566 | |||||||||||
| CAZy Family | CBM32 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 28772; End: 32239 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CBM32 | 108 | 237 | 4.2e-18 | 0.8548387096774194 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00754 | F5_F8_type_C | 5.18e-16 | 108 | 236 | 12 | 127 | F5/8 type C domain. This domain is also known as the discoidin (DS) domain family. |
| cd14791 | GH36 | 6.89e-10 | 450 | 553 | 1 | 87 | glycosyl hydrolase family 36 (GH36). GH36 enzymes occur in prokaryotes, eukaryotes, and archaea with a wide range of hydrolytic activities, including alpha-galactosidase, alpha-N-acetylgalactosaminidase, stachyose synthase, and raffinose synthase. All GH36 enzymes cleave a terminal carbohydrate moiety from a substrate that varies considerably in size, depending on the enzyme, and may be either a starch or a glycoprotein. GH36 members are retaining enzymes that cleave their substrates via an acid/base-catalyzed, double-displacement mechanism involving a covalent glycosyl-enzyme intermediate. Two aspartic acid residues have been identified as the catalytic nucleophile and the acid/base, respectively. |
| cd00057 | FA58C | 5.51e-05 | 97 | 230 | 16 | 137 | Substituted updates: Jan 31, 2002 |
| COG3345 | GalA | 1.47e-04 | 457 | 553 | 294 | 377 | Alpha-galactosidase [Carbohydrate transport and metabolism]. |
| cd00110 | LamG | 4.32e-04 | 1063 | 1116 | 44 | 100 | Laminin G domain; Laminin G-like domains are usually Ca++ mediated receptors that can have binding sites for steroids, beta1 integrins, heparin, sulfatides, fibulin-1, and alpha-dystroglycans. Proteins that contain LamG domains serve a variety of purposes including signal transduction via cell-surface steroid receptors, adhesion, migration and differentiation through mediation of cell adhesion molecules. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QUT26415.1 | 9.53e-230 | 245 | 1140 | 86 | 930 |
| QDO70357.1 | 3.85e-227 | 253 | 1137 | 94 | 927 |
| AXH52486.1 | 7.40e-199 | 25 | 1141 | 39 | 1184 |
| SQI04569.1 | 1.45e-198 | 25 | 1141 | 39 | 1184 |
| AOY54034.1 | 2.83e-198 | 14 | 1141 | 27 | 1184 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4A41_A | 2.87e-11 | 87 | 235 | 26 | 156 | CpGH89CBM32-5,from Clostridium perfringens, in complex with galactose [Clostridium perfringens],4A44_A CpGH89CBM32-5, from Clostridium perfringens, in complex with the Tn Antigen [Clostridium perfringens],4A45_A CpGH89CBM32-5, from Clostridium perfringens, in complex with GalNAc- beta-1,3-galactose [Clostridium perfringens],4AAX_A CpGH89CBM32-5, from Clostridium perfringens, in complex with N- acetylgalactosamine [Clostridium perfringens] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
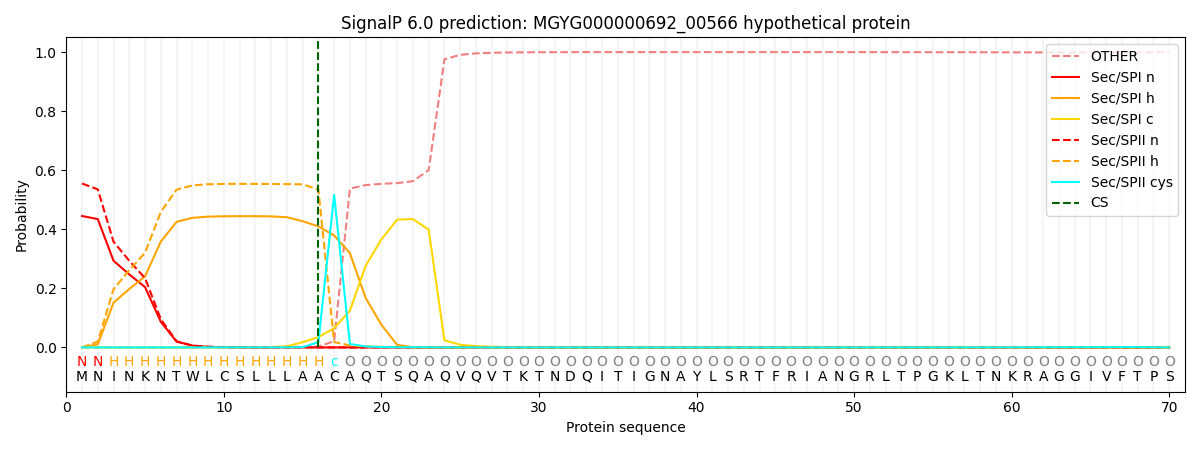
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000358 | 0.437055 | 0.561997 | 0.000189 | 0.000208 | 0.000178 |
