You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000693_01499
Basic Information
help
| Species |
|
| Lineage |
Bacteria; Actinobacteriota; Coriobacteriia; Coriobacteriales; Coriobacteriaceae; Collinsella;
|
| CAZyme ID |
MGYG000000693_01499
|
| CAZy Family |
GT83 |
| CAZyme Description |
hypothetical protein
|
| CAZyme Property |
| Protein Length |
CGC |
Molecular Weight |
Isoelectric Point |
| 713 |
|
74243.81 |
8.3399 |
|
| Genome Property |
| Genome Assembly ID |
Genome Size |
Genome Type |
Country |
Continent |
| MGYG000000693 |
2022975 |
MAG |
Kazakhstan |
Asia |
|
| Gene Location |
Start: 6225;
End: 8366
Strand: +
|
No EC number prediction in MGYG000000693_01499.
| Family |
Start |
End |
Evalue |
family coverage |
| GT83 |
38 |
528 |
1.7e-38 |
0.7833333333333333 |
| Cdd ID |
Domain |
E-Value |
qStart |
qEnd |
sStart |
sEnd |
Domain Description |
| COG1807
|
ArnT |
1.77e-23 |
79 |
532 |
46 |
408 |
4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231
|
PMT_2 |
3.37e-23 |
96 |
259 |
1 |
152 |
Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG5305
|
COG5305 |
0.005 |
96 |
257 |
96 |
258 |
Uncharacterized membrane protein [Function unknown]. |
| Hit ID |
E-Value |
Query Start |
Query End |
Hit Start |
Hit End |
Description |
|
O34575
|
8.19e-63 |
41 |
696 |
10 |
660 |
Putative mannosyltransferase YkcB OS=Bacillus subtilis (strain 168) OX=224308 GN=ykcB PE=3 SV=2 |
|
P37483
|
2.71e-52 |
42 |
696 |
9 |
647 |
Putative mannosyltransferase YycA OS=Bacillus subtilis (strain 168) OX=224308 GN=yycA PE=3 SV=2 |
This protein is predicted as OTHER
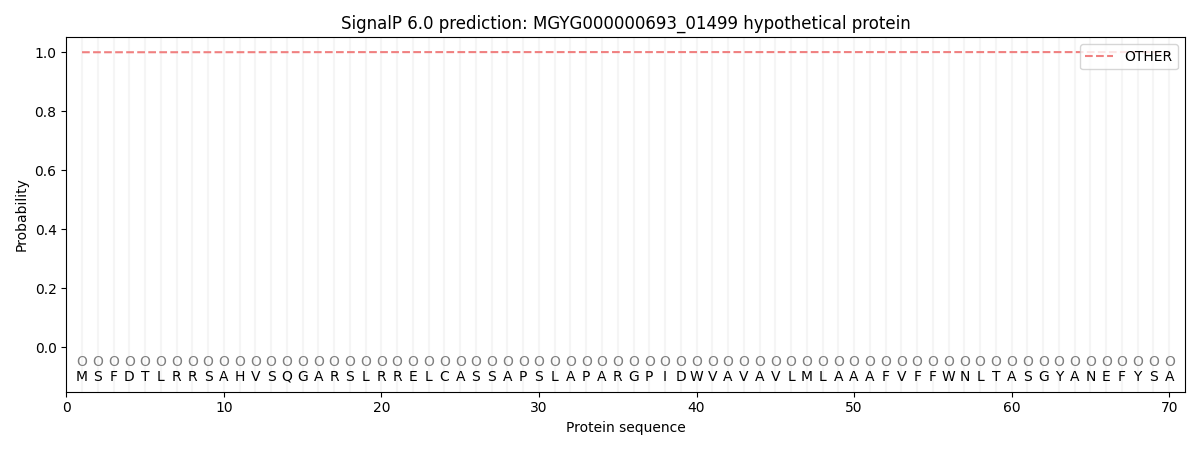
| Other |
SP_Sec_SPI |
LIPO_Sec_SPII |
TAT_Tat_SPI |
TATLIP_Sec_SPII |
PILIN_Sec_SPIII |
|
0.999786
|
0.000236
|
0.000000
|
0.000000
|
0.000000
|
0.000000
|
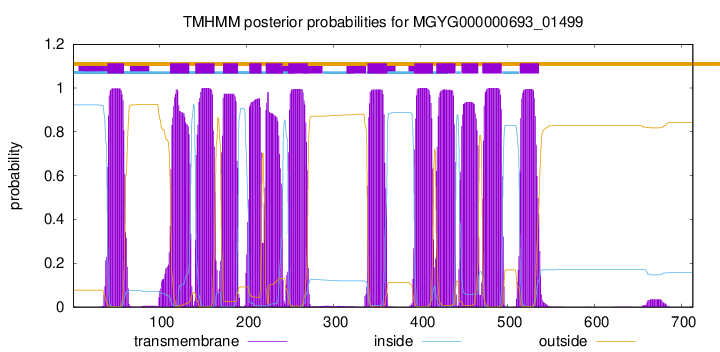
| start |
end |
| 40 |
59 |
| 112 |
134 |
| 141 |
163 |
| 173 |
190 |
| 203 |
217 |
| 222 |
241 |
| 248 |
270 |
| 339 |
361 |
| 393 |
414 |
| 418 |
440 |
| 447 |
466 |
| 471 |
493 |
| 514 |
536 |


