You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000695_00033
You are here: Home > Sequence: MGYG000000695_00033
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp000436035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000436035 | |||||||||||
| CAZyme ID | MGYG000000695_00033 | |||||||||||
| CAZy Family | GH106 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 38097; End: 40694 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH106 | 27 | 576 | 1.7e-169 | 0.6092233009708737 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam17132 | Glyco_hydro_106 | 3.44e-28 | 196 | 571 | 392 | 755 | alpha-L-rhamnosidase. |
| COG3250 | LacZ | 2.19e-04 | 700 | 812 | 11 | 133 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCS85006.1 | 0.0 | 19 | 858 | 12 | 834 |
| ADE82941.1 | 0.0 | 22 | 864 | 16 | 804 |
| QVJ80546.1 | 0.0 | 22 | 864 | 16 | 804 |
| QJR98068.1 | 0.0 | 35 | 862 | 6 | 859 |
| AGB29019.1 | 0.0 | 13 | 862 | 8 | 854 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5MQM_A | 6.99e-29 | 197 | 822 | 412 | 1065 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron],5MQN_A Glycoside hydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 5MWK_A | 1.60e-28 | 197 | 822 | 412 | 1065 | Glycosidehydrolase BT_0986 [Bacteroides thetaiotaomicron] |
| 6Q2F_A | 6.08e-22 | 196 | 863 | 455 | 1139 | Structureof Rhamnosidase from Novosphingobium sp. PP1Y [Novosphingobium sp. PP1Y] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
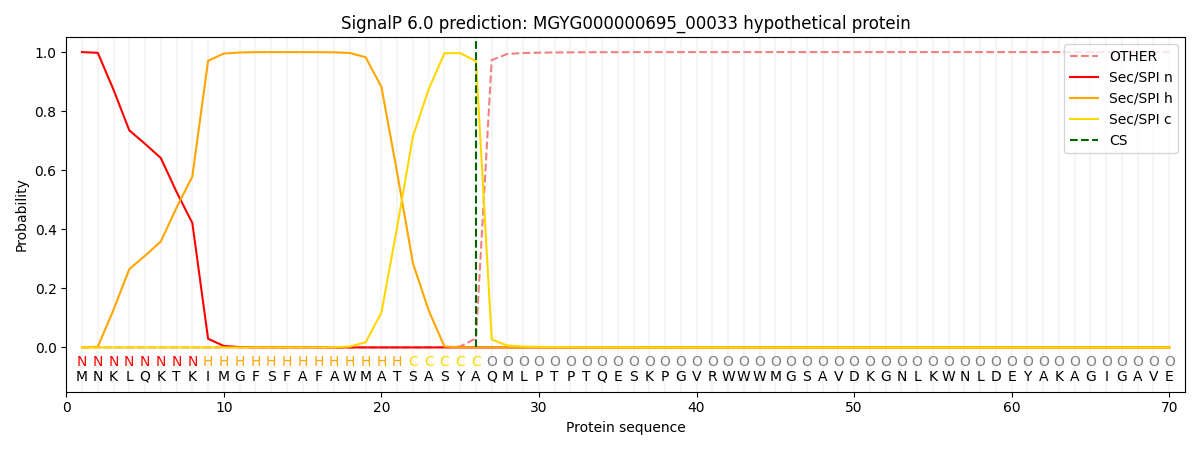
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000446 | 0.998812 | 0.000179 | 0.000199 | 0.000181 | 0.000162 |
