You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000695_00917
You are here: Home > Sequence: MGYG000000695_00917
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Prevotella sp000436035 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; Prevotella; Prevotella sp000436035 | |||||||||||
| CAZyme ID | MGYG000000695_00917 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 2175; End: 4403 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 465 | 727 | 3.2e-41 | 0.6765676567656765 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 3.79e-25 | 466 | 727 | 54 | 263 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 4.71e-24 | 463 | 728 | 93 | 309 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 1.37e-17 | 463 | 708 | 116 | 311 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam00331 | Glyco_hydro_10 | 4.08e-10 | 56 | 141 | 1 | 85 | Glycosyl hydrolase family 10. |
| smart00633 | Glyco_10 | 4.59e-05 | 100 | 142 | 1 | 43 | Glycosyl hydrolase family 10. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNT67590.1 | 0.0 | 1 | 742 | 1 | 734 |
| QIU93949.1 | 1.92e-258 | 10 | 731 | 10 | 725 |
| QDM11106.1 | 1.06e-257 | 10 | 731 | 10 | 725 |
| QUR44907.1 | 2.21e-257 | 10 | 731 | 10 | 725 |
| QGT73069.1 | 3.13e-257 | 10 | 731 | 10 | 725 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MGQ_A | 6.79e-17 | 146 | 308 | 4 | 155 | PbXyn10CCBM APO [Prevotella bryantii B14] |
| 4W8L_A | 2.18e-13 | 467 | 666 | 106 | 281 | Structureof GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_B Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis],4W8L_C Structure of GH10 from Paenibacillus barcinonensis [Paenibacillus barcinonensis] |
| 1W32_A | 1.21e-10 | 478 | 698 | 109 | 310 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W32_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1W3H_A | 1.29e-10 | 478 | 698 | 120 | 321 | The3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus],1W3H_B The 3-dimensional structure of a thermostable mutant of a xylanase (Xyn10A) from Cellvibrio japonicus [Cellvibrio japonicus] |
| 1CLX_A | 1.60e-10 | 478 | 698 | 108 | 309 | CatalyticCore Of Xylanase A [Cellvibrio japonicus],1CLX_B Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_C Catalytic Core Of Xylanase A [Cellvibrio japonicus],1CLX_D Catalytic Core Of Xylanase A [Cellvibrio japonicus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O69230 | 4.58e-12 | 467 | 666 | 472 | 647 | Endo-1,4-beta-xylanase C OS=Paenibacillus barcinonensis OX=198119 GN=xynC PE=1 SV=1 |
| P14768 | 1.87e-09 | 478 | 698 | 372 | 573 | Endo-1,4-beta-xylanase A OS=Cellvibrio japonicus (strain Ueda107) OX=498211 GN=xynA PE=1 SV=2 |
| P07986 | 1.40e-08 | 486 | 733 | 157 | 355 | Exoglucanase/xylanase OS=Cellulomonas fimi OX=1708 GN=cex PE=1 SV=1 |
| Q59675 | 9.52e-06 | 65 | 140 | 258 | 338 | Endo-beta-1,4-xylanase Xyn10C OS=Cellvibrio japonicus OX=155077 GN=xyn10C PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
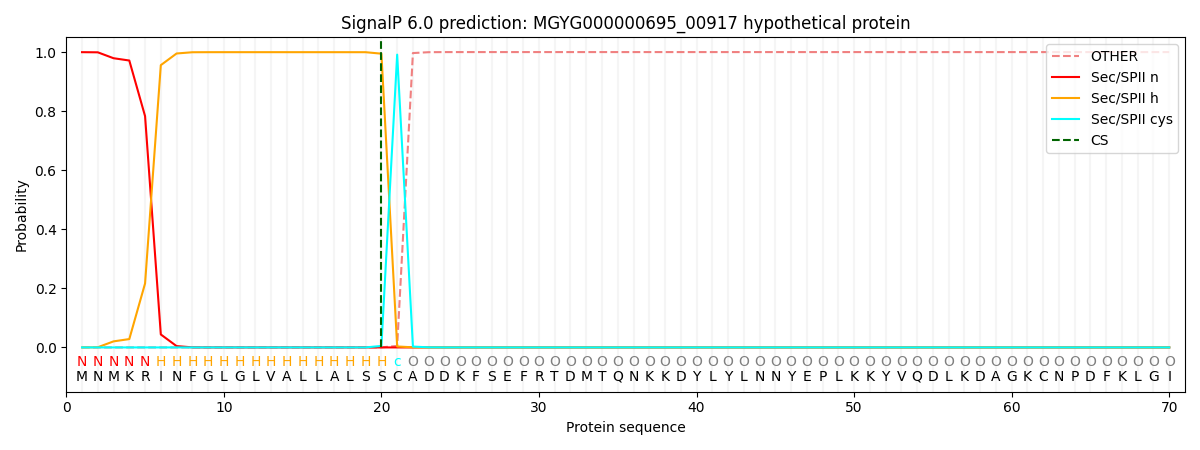
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000276 | 0.999774 | 0.000000 | 0.000000 | 0.000000 |
