You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000700_01091
You are here: Home > Sequence: MGYG000000700_01091
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA4372 sp900543815 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; Bacteroidaceae; UBA4372; UBA4372 sp900543815 | |||||||||||
| CAZyme ID | MGYG000000700_01091 | |||||||||||
| CAZy Family | GH10 | |||||||||||
| CAZyme Description | Endo-1,4-beta-xylanase/feruloyl esterase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3768; End: 4919 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH10 | 52 | 378 | 1.9e-103 | 0.9900990099009901 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| smart00633 | Glyco_10 | 2.96e-111 | 93 | 374 | 1 | 261 | Glycosyl hydrolase family 10. |
| pfam00331 | Glyco_hydro_10 | 2.46e-108 | 52 | 378 | 1 | 310 | Glycosyl hydrolase family 10. |
| COG3693 | XynA | 7.29e-92 | 77 | 378 | 51 | 339 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QQZ02686.1 | 3.40e-149 | 44 | 382 | 30 | 374 |
| AGC54684.1 | 1.58e-141 | 51 | 378 | 33 | 369 |
| QVJ82151.1 | 8.89e-136 | 31 | 378 | 4 | 368 |
| ACN78954.1 | 1.26e-135 | 31 | 378 | 4 | 368 |
| ADE83639.1 | 1.26e-135 | 31 | 378 | 4 | 368 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5AY7_A | 1.94e-125 | 50 | 378 | 17 | 345 | Apsychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5AY7_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [Aegilops speltoides subsp. speltoides],5D4Y_A A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples],5D4Y_B A psychrophilic glycoside hydrolase family 10 endo-beta-1,4-xylanase [environmental samples] |
| 6WQW_A | 7.76e-91 | 52 | 381 | 6 | 331 | ChainA, Beta-xylanase [Thermobacillus composti] |
| 1N82_A | 1.02e-87 | 52 | 374 | 8 | 326 | Thehigh-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],1N82_B The high-resolution crystal structure of IXT6, a thermophilic, intracellular xylanase from G. stearothermophilus [Geobacillus stearothermophilus],3MUA_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUA_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 2Q8X_A | 1.02e-87 | 52 | 374 | 8 | 326 | Thehigh-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],2Q8X_B The high-resolution crystal structure of ixt6, a thermophilic, intracellular xylanase from G. stearothermophilus [unidentified],3MSD_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSD_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_A Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MSG_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
| 3MUI_A | 8.19e-87 | 52 | 374 | 8 | 326 | Enzyme-Substrateinteractions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus],3MUI_B Enzyme-Substrate interactions of IXT6, the intracellular xylanase of G. stearothermophilus. [Geobacillus stearothermophilus] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| D5EY13 | 2.51e-136 | 31 | 378 | 4 | 368 | Endo-1,4-beta-xylanase/feruloyl esterase OS=Prevotella ruminicola (strain ATCC 19189 / JCM 8958 / 23) OX=264731 GN=xyn10D-fae1A PE=1 SV=1 |
| O69231 | 3.70e-85 | 49 | 374 | 5 | 326 | Endo-1,4-beta-xylanase B OS=Paenibacillus barcinonensis OX=198119 GN=xynB PE=1 SV=1 |
| Q12603 | 9.78e-82 | 44 | 374 | 27 | 347 | Beta-1,4-xylanase OS=Dictyoglomus thermophilum OX=14 GN=xynA PE=3 SV=1 |
| P10474 | 1.61e-80 | 49 | 381 | 42 | 374 | Endoglucanase/exoglucanase B OS=Caldicellulosiruptor saccharolyticus OX=44001 GN=celB PE=3 SV=1 |
| P45703 | 4.49e-77 | 47 | 376 | 3 | 327 | Endo-1,4-beta-xylanase OS=Geobacillus stearothermophilus OX=1422 GN=xynA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
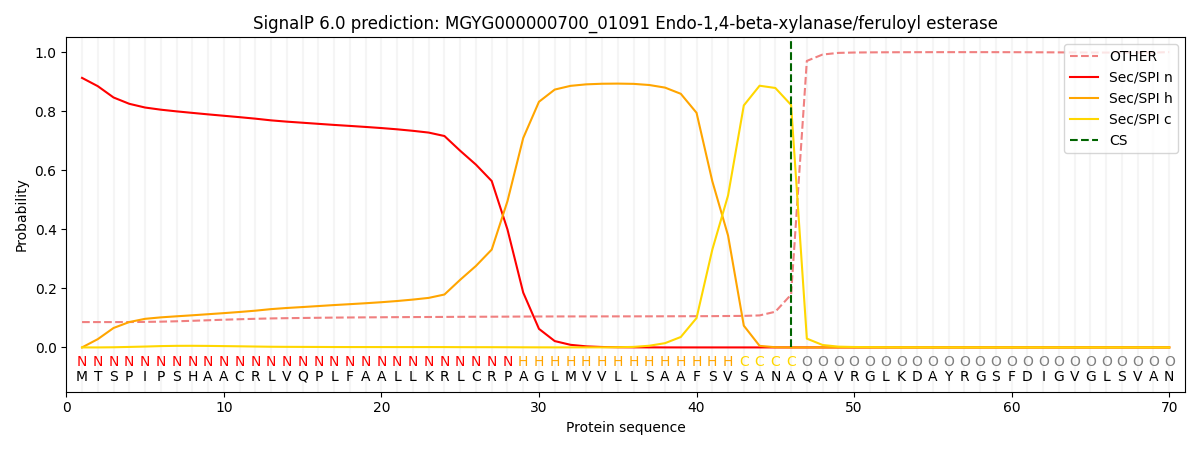
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.089632 | 0.906731 | 0.002431 | 0.000423 | 0.000372 | 0.000395 |
