You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000710_00773
You are here: Home > Sequence: MGYG000000710_00773
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; CAG-110; | |||||||||||
| CAZyme ID | MGYG000000710_00773 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | Beta-galactosidase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 11172; End: 14372 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 204 | 648 | 5.6e-83 | 0.5093085106382979 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 9.18e-42 | 214 | 691 | 47 | 498 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| PRK10340 | ebgA | 1.27e-36 | 209 | 647 | 71 | 514 | cryptic beta-D-galactosidase subunit alpha; Reviewed |
| PRK10150 | PRK10150 | 9.51e-30 | 184 | 648 | 16 | 512 | beta-D-glucuronidase; Provisional |
| PRK09525 | lacZ | 1.82e-19 | 241 | 647 | 124 | 540 | beta-galactosidase. |
| pfam02837 | Glyco_hydro_2_N | 2.83e-17 | 231 | 344 | 59 | 165 | Glycosyl hydrolases family 2, sugar binding domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities and has a jelly-roll fold. The domain binds the sugar moiety during the sugar-hydrolysis reaction. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QNK55911.1 | 1.10e-178 | 135 | 1064 | 219 | 1234 |
| QTH41860.1 | 1.18e-53 | 213 | 1027 | 23 | 858 |
| SDS96723.1 | 2.07e-51 | 237 | 1062 | 44 | 923 |
| ACZ41351.1 | 2.86e-43 | 235 | 1038 | 48 | 904 |
| QNK56508.1 | 1.32e-39 | 235 | 932 | 55 | 780 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5T98_A | 9.11e-25 | 207 | 600 | 52 | 448 | Crystalstructure of BuGH2Awt [Bacteroides uniformis],5T98_B Crystal structure of BuGH2Awt [Bacteroides uniformis],5T99_A Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis],5T99_B Crystal structure of BuGH2Awt in complex with Galactoisofagomine [Bacteroides uniformis] |
| 6D8K_A | 1.81e-24 | 235 | 599 | 96 | 440 | Bacteroidesmultiple species beta-glucuronidase [Bacteroides ovatus],6D8K_B Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_C Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus],6D8K_D Bacteroides multiple species beta-glucuronidase [Bacteroides ovatus] |
| 3FN9_A | 2.28e-22 | 235 | 606 | 58 | 433 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 1JZ7_A | 2.78e-22 | 207 | 647 | 71 | 539 | E.COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_B E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_C E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],1JZ7_D E. COLI (lacZ) BETA-GALACTOSIDASE IN COMPLEX WITH GALACTOSE [Escherichia coli],4TTG_A Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_B Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_C Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli],4TTG_D Beta-galactosidase (E. coli) in the presence of potassium chloride. [Escherichia coli] |
| 3CZJ_A | 2.78e-22 | 207 | 647 | 71 | 539 | ChainA, Beta-galactosidase [Escherichia coli K-12],3CZJ_B Chain B, Beta-galactosidase [Escherichia coli K-12],3CZJ_C Chain C, Beta-galactosidase [Escherichia coli K-12],3CZJ_D Chain D, Beta-galactosidase [Escherichia coli K-12] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| T2KM09 | 1.12e-23 | 237 | 607 | 106 | 462 | Putative beta-glucuronidase OS=Formosa agariphila (strain DSM 15362 / KCTC 12365 / LMG 23005 / KMM 3901 / M-2Alg 35-1) OX=1347342 GN=BN863_22050 PE=2 SV=2 |
| B4S2K9 | 2.99e-22 | 241 | 647 | 124 | 538 | Beta-galactosidase OS=Alteromonas mediterranea (strain DSM 17117 / CIP 110805 / LMG 28347 / Deep ecotype) OX=1774373 GN=lacZ PE=3 SV=1 |
| Q8X685 | 6.71e-22 | 207 | 647 | 72 | 540 | Beta-galactosidase OS=Escherichia coli O157:H7 OX=83334 GN=lacZ PE=3 SV=1 |
| B5Z2P7 | 6.71e-22 | 207 | 647 | 72 | 540 | Beta-galactosidase OS=Escherichia coli O157:H7 (strain EC4115 / EHEC) OX=444450 GN=lacZ PE=3 SV=1 |
| Q32JB6 | 1.16e-21 | 231 | 647 | 114 | 540 | Beta-galactosidase OS=Shigella dysenteriae serotype 1 (strain Sd197) OX=300267 GN=lacZ PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as LIPO
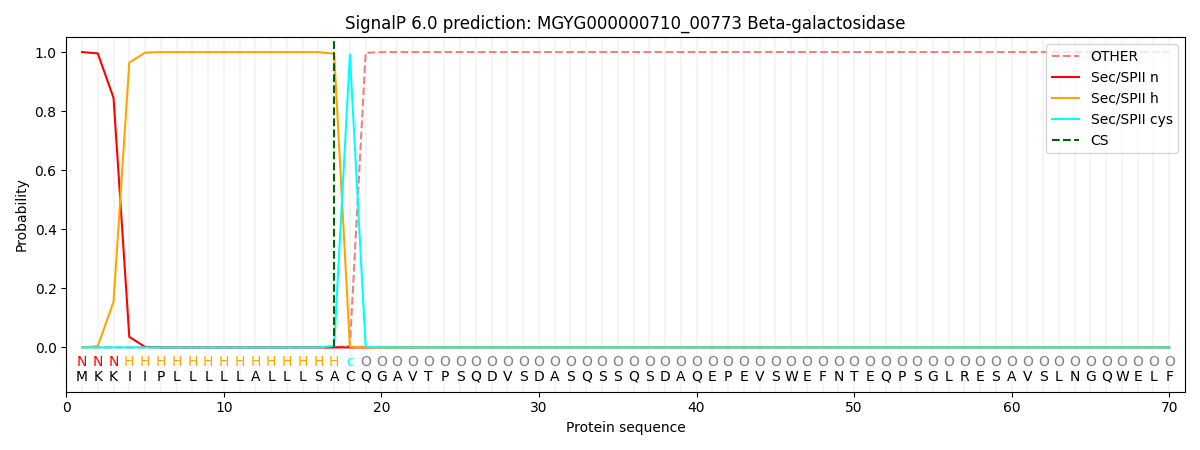
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000000 | 0.000002 | 1.000058 | 0.000000 | 0.000000 | 0.000000 |
