You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000712_01877
You are here: Home > Sequence: MGYG000000712_01877
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | Fusobacterium_B sp900541465 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Fusobacteriota; Fusobacteriia; Fusobacteriales; Fusobacteriaceae; Fusobacterium_B; Fusobacterium_B sp900541465 | |||||||||||
| CAZyme ID | MGYG000000712_01877 | |||||||||||
| CAZy Family | GT83 | |||||||||||
| CAZyme Description | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3; End: 1505 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GT83 | 14 | 368 | 6e-55 | 0.6407407407407407 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG1807 | ArnT | 3.96e-20 | 35 | 335 | 30 | 325 | 4-amino-4-deoxy-L-arabinose transferase or related glycosyltransferase of PMT family [Cell wall/membrane/envelope biogenesis]. |
| pfam13231 | PMT_2 | 4.80e-08 | 68 | 231 | 1 | 159 | Dolichyl-phosphate-mannose-protein mannosyltransferase. This family contains members that are not captured by pfam02366. |
| COG1287 | Stt3 | 0.004 | 68 | 228 | 86 | 253 | Asparagine N-glycosylation enzyme, membrane subunit Stt3 [Posttranslational modification, protein turnover, chaperones]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVQ28191.1 | 1.04e-154 | 13 | 495 | 8 | 489 |
| SQI99970.1 | 1.04e-154 | 13 | 495 | 8 | 489 |
| AVQ30207.1 | 1.51e-143 | 26 | 495 | 21 | 488 |
| BBA52631.1 | 4.89e-142 | 11 | 495 | 5 | 488 |
| AVQ18501.1 | 3.96e-138 | 13 | 496 | 9 | 487 |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| O67270 | 2.86e-12 | 22 | 353 | 5 | 328 | Uncharacterized protein aq_1220 OS=Aquifex aeolicus (strain VF5) OX=224324 GN=aq_1220 PE=3 SV=1 |
| Q8D339 | 2.73e-09 | 13 | 387 | 13 | 397 | Undecaprenyl phosphate-alpha-4-amino-4-deoxy-L-arabinose arabinosyl transferase OS=Wigglesworthia glossinidia brevipalpis OX=36870 GN=arnT PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as OTHER
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.999975 | 0.000032 | 0.000001 | 0.000000 | 0.000000 | 0.000026 |
TMHMM Annotations download full data without filtering help
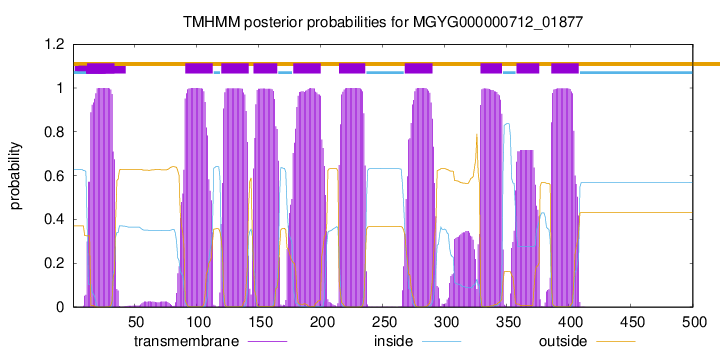
| start | end |
|---|---|
| 12 | 34 |
| 91 | 113 |
| 120 | 142 |
| 146 | 165 |
| 178 | 200 |
| 215 | 236 |
| 268 | 290 |
| 329 | 346 |
| 358 | 376 |
| 386 | 408 |
