You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_00174
You are here: Home > Sequence: MGYG000000720_00174
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_00174 | |||||||||||
| CAZy Family | GH110 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 65271; End: 68834 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH110 | 195 | 737 | 3.8e-46 | 0.9872262773722628 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam13472 | Lipase_GDSL_2 | 7.71e-14 | 994 | 1174 | 3 | 175 | GDSL-like Lipase/Acylhydrolase family. This family of presumed lipases and related enzymes are similar to pfam00657. |
| pfam13229 | Beta_helix | 1.62e-12 | 573 | 767 | 6 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| cd00229 | SGNH_hydrolase | 5.95e-11 | 990 | 1183 | 1 | 187 | SGNH_hydrolase, or GDSL_hydrolase, is a diverse family of lipases and esterases. The tertiary fold of the enzyme is substantially different from that of the alpha/beta hydrolase family and unique among all known hydrolases; its active site closely resembles the typical Ser-His-Asp(Glu) triad from other serine hydrolases, but may lack the carboxlic acid. |
| pfam13229 | Beta_helix | 1.77e-10 | 571 | 708 | 50 | 156 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
| COG2755 | TesA | 4.96e-07 | 989 | 1182 | 10 | 206 | Lysophospholipase L1 or related esterase [Amino acid transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46987.1 | 0.0 | 25 | 1001 | 1 | 976 |
| AVM44429.1 | 4.09e-127 | 1 | 771 | 1 | 769 |
| AVM44109.1 | 1.19e-121 | 36 | 771 | 81 | 806 |
| AVM44539.1 | 4.50e-118 | 37 | 771 | 42 | 764 |
| QTH44371.1 | 3.28e-80 | 206 | 712 | 46 | 529 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 7JW4_A | 6.35e-18 | 181 | 710 | 17 | 566 | Crystalstructure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta],7JW4_B Crystal structure of PdGH110B in complex with D-galactose [Pseudoalteromonas distincta] |
| 7JWF_A | 2.53e-17 | 181 | 710 | 17 | 566 | Crystalstructure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_B Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_C Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta],7JWF_D Crystal structure of PdGH110B D344N in complex with alpha-(1,3)-galactobiose [Pseudoalteromonas distincta] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B1KD83 | 1.02e-16 | 196 | 739 | 37 | 580 | Alpha-1,3-galactosidase A OS=Shewanella woodyi (strain ATCC 51908 / MS32) OX=392500 GN=glaA PE=3 SV=1 |
| B2UNU8 | 2.99e-13 | 191 | 711 | 218 | 741 | Alpha-1,3-galactosidase B OS=Akkermansia muciniphila (strain ATCC BAA-835 / DSM 22959 / JCM 33894 / BCRC 81048 / CCUG 64013 / CIP 107961 / Muc) OX=349741 GN=glaB PE=3 SV=1 |
| B1V8K7 | 3.43e-10 | 360 | 715 | 219 | 589 | Alpha-1,3-galactosidase A OS=Streptacidiphilus griseoplanus OX=66896 GN=glaA PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
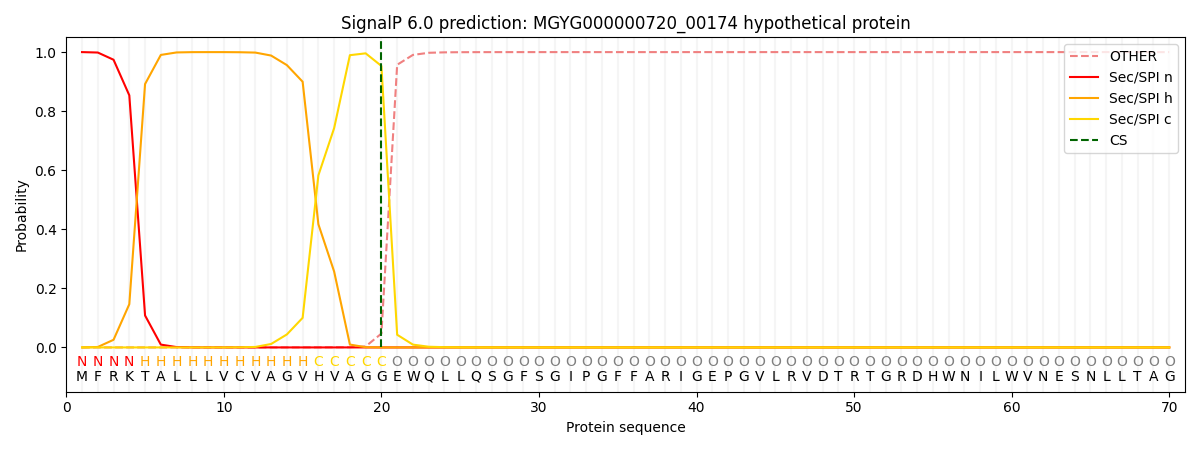
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000551 | 0.998685 | 0.000205 | 0.000185 | 0.000171 | 0.000173 |
