You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_00751
You are here: Home > Sequence: MGYG000000720_00751
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_00751 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 54490; End: 56007 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH5 | 199 | 470 | 1.7e-41 | 0.7272727272727273 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam00150 | Cellulase | 2.70e-19 | 206 | 464 | 35 | 272 | Cellulase (glycosyl hydrolase family 5). |
| COG2730 | BglC | 3.22e-17 | 200 | 478 | 78 | 377 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM45719.1 | 1.55e-178 | 30 | 501 | 77 | 549 |
| AVM46198.1 | 3.22e-170 | 28 | 503 | 29 | 501 |
| AVM45721.1 | 3.16e-169 | 3 | 502 | 11 | 514 |
| AVM46390.1 | 3.55e-160 | 2 | 505 | 5 | 505 |
| AVM45720.1 | 5.50e-158 | 18 | 503 | 79 | 563 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6KDD_A | 2.20e-13 | 202 | 465 | 48 | 295 | endoglucanase[Fervidobacterium pennivorans DSM 9078] |
| 4LX4_A | 2.53e-10 | 257 | 455 | 94 | 275 | CrystalStructure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_B Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_C Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],4LX4_D Crystal Structure Determination of Pseudomonas stutzeri endoglucanase Cel5A using a Twinned Data Set [Pseudomonas stutzeri A1501],6R2J_A Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_B Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_C Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501],6R2J_D Crystal Structure of Pseudomonas stutzeri endoglucanase Cel5A in complex with cellobiose [Pseudomonas stutzeri A1501] |
| 1ECE_A | 9.35e-10 | 201 | 483 | 50 | 331 | AcidothermusCellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus],1ECE_B Acidothermus Cellulolyticus Endocellulase E1 Catalytic Domain In Complex With A Cellotetraose [Acidothermus cellulolyticus] |
| 1VRX_A | 5.20e-09 | 201 | 483 | 50 | 331 | ChainA, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus],1VRX_B Chain B, ENDOCELLULASE E1 FROM A. CELLULOLYTICUS [Acidothermus cellulolyticus] |
| 4EE9_A | 3.22e-08 | 199 | 434 | 33 | 247 | Crystalstructure of the RBcel1 endo-1,4-glucanase [uncultured bacterium],4M24_A Crystal structure of the endo-1,4-glucanase, RBcel1, in complex with cellobiose [uncultured bacterium] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P16169 | 1.90e-10 | 200 | 393 | 35 | 225 | Cellodextrinase A OS=Ruminococcus flavefaciens OX=1265 GN=celA PE=3 SV=3 |
| W8QRE4 | 5.90e-10 | 196 | 436 | 71 | 296 | Beta-xylosidase OS=Phanerodontia chrysosporium OX=2822231 GN=Xyl5 PE=1 SV=2 |
| P54583 | 8.50e-09 | 201 | 483 | 91 | 372 | Endoglucanase E1 OS=Acidothermus cellulolyticus (strain ATCC 43068 / DSM 8971 / 11B) OX=351607 GN=Acel_0614 PE=1 SV=1 |
| P23548 | 1.06e-08 | 201 | 434 | 83 | 319 | Endoglucanase OS=Paenibacillus polymyxa OX=1406 PE=3 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
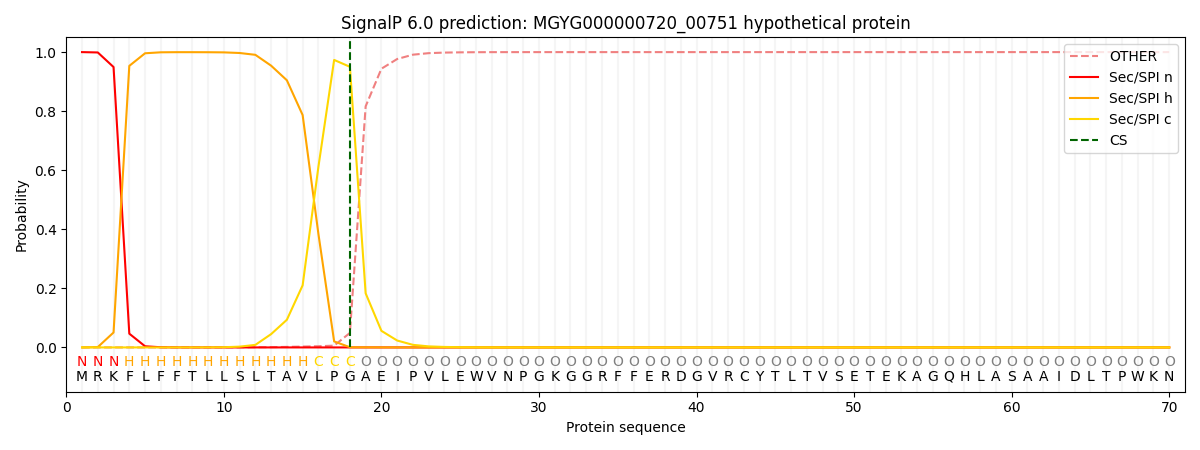
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000649 | 0.998531 | 0.000305 | 0.000185 | 0.000165 | 0.000163 |
