You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_01168
You are here: Home > Sequence: MGYG000000720_01168
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_01168 | |||||||||||
| CAZy Family | GH39 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 30278; End: 32152 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH39 | 75 | 239 | 1.9e-28 | 0.4709976798143852 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3693 | XynA | 2.96e-07 | 66 | 171 | 69 | 185 | Endo-1,4-beta-xylanase, GH35 family [Carbohydrate transport and metabolism]. |
| pfam01229 | Glyco_hydro_39 | 4.40e-07 | 54 | 243 | 45 | 276 | Glycosyl hydrolases family 39. |
| COG2723 | BglB | 5.40e-05 | 52 | 217 | 68 | 240 | Beta-glucosidase/6-phospho-beta-glucosidase/beta-galactosidase [Carbohydrate transport and metabolism]. |
| COG1874 | GanA | 2.46e-04 | 31 | 170 | 18 | 190 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| COG2730 | BglC | 4.32e-04 | 38 | 226 | 68 | 271 | Aryl-phospho-beta-D-glucosidase BglC, GH1 family [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44885.1 | 8.06e-193 | 10 | 618 | 11 | 618 |
| QDU57372.1 | 7.15e-115 | 12 | 616 | 11 | 630 |
| AVM44901.1 | 9.13e-77 | 1 | 578 | 1 | 597 |
| AHF90941.1 | 2.71e-57 | 16 | 576 | 216 | 823 |
| QAT16636.1 | 1.18e-32 | 52 | 241 | 126 | 325 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4ZN2_A | 1.80e-11 | 51 | 255 | 41 | 266 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 1.80e-11 | 51 | 255 | 41 | 266 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
| 5JVK_A | 4.76e-11 | 32 | 208 | 107 | 290 | Structuralinsights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_B Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus],5JVK_C Structural insights into a family 39 glycoside hydrolase from the gut symbiont Bacteroides cellulosilyticus WH2. [Bacteroides cellulosilyticus] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
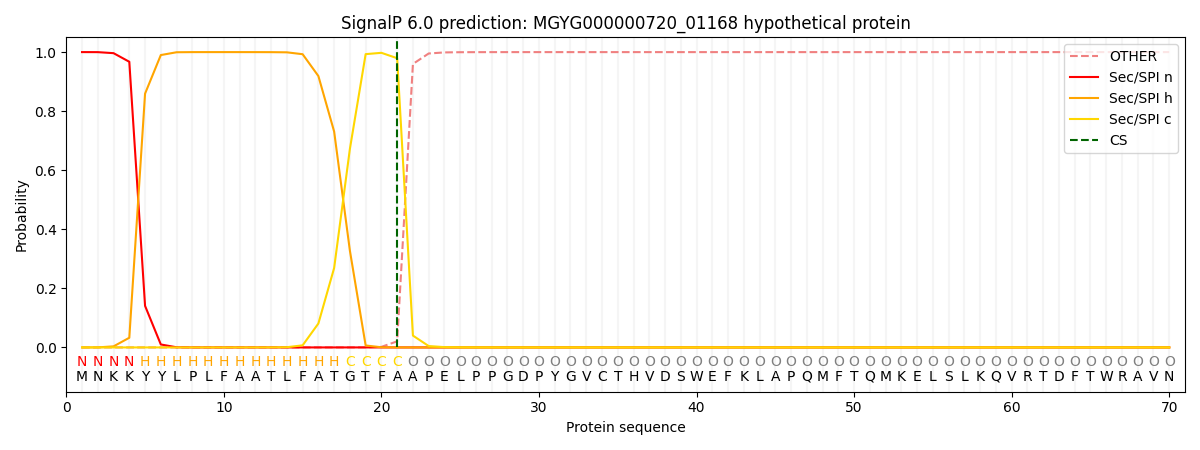
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000274 | 0.999062 | 0.000170 | 0.000162 | 0.000153 | 0.000141 |
