You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_01867
You are here: Home > Sequence: MGYG000000720_01867
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_01867 | |||||||||||
| CAZy Family | GH42 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 742; End: 4827 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH42 | 182 | 352 | 8.9e-19 | 0.4636118598382749 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam02449 | Glyco_hydro_42 | 1.03e-11 | 195 | 486 | 100 | 375 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 3.69e-09 | 99 | 377 | 35 | 317 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM45830.1 | 0.0 | 3 | 1361 | 2 | 1351 |
| AVM46758.1 | 1.33e-68 | 37 | 1023 | 184 | 1150 |
| AVM46955.1 | 1.99e-27 | 835 | 1031 | 1011 | 1214 |
| AVM45112.1 | 2.99e-26 | 820 | 1021 | 993 | 1190 |
| AVM45809.1 | 3.02e-24 | 804 | 1074 | 983 | 1255 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 6PTM_A | 1.51e-11 | 193 | 664 | 253 | 722 | Crystalstructure of apo exo-carrageenase GH42 from Bacteroides ovatus [Bacteroides ovatus CL02T12C04] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q93GI5 | 1.01e-10 | 95 | 702 | 32 | 625 | Beta-galactosidase III OS=Bifidobacterium longum subsp. infantis OX=1682 GN=beta-galIII PE=1 SV=1 |
| C8WV58 | 2.99e-10 | 119 | 705 | 54 | 622 | Beta-galactosidase BglY OS=Alicyclobacillus acidocaldarius subsp. acidocaldarius (strain ATCC 27009 / DSM 446 / BCRC 14685 / JCM 5260 / KCTC 1825 / NBRC 15652 / NCIMB 11725 / NRRL B-14509 / 104-IA) OX=521098 GN=bglY PE=1 SV=1 |
| C9S0R2 | 3.87e-10 | 170 | 745 | 89 | 634 | Beta-galactosidase BgaB OS=Geobacillus sp. (strain Y412MC61) OX=544556 GN=bgaB PE=3 SV=1 |
| P19668 | 5.08e-10 | 170 | 745 | 89 | 634 | Beta-galactosidase bgaB OS=Geobacillus kaustophilus OX=1462 GN=bgaB PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
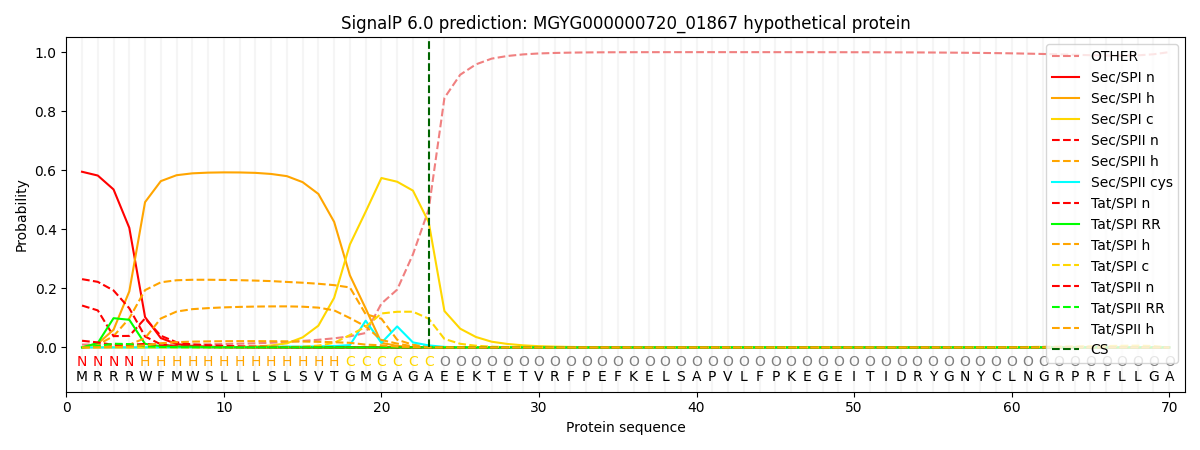
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.014225 | 0.578362 | 0.235843 | 0.146234 | 0.024589 | 0.000725 |
