You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_01935
You are here: Home > Sequence: MGYG000000720_01935
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
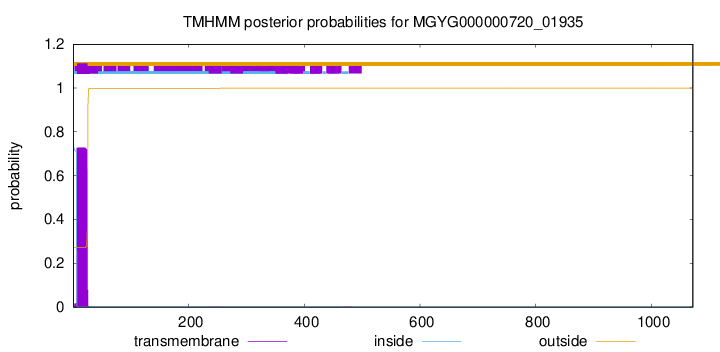
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_01935 | |||||||||||
| CAZy Family | GH5 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 23259; End: 26477 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH167 | 334 | 871 | 3.1e-43 | 0.7684797768479776 |
| GH5 | 70 | 345 | 1.4e-18 | 0.7672727272727272 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| cd03143 | A4_beta-galactosidase_middle_domain | 8.48e-10 | 771 | 884 | 3 | 113 | A4 beta-galactosidase middle domain: a type 1 glutamine amidotransferase (GATase1)-like domain. A4 beta-galactosidase middle domain: a type 1 glutamine amidotransferase (GATase1)-like domain. This group includes proteins similar to beta-galactosidase from Thermus thermophilus. Beta-Galactosidase hydrolyzes the beta-1,4-D-galactosidic linkage of lactose, as well as those of related chromogens, o-nitrophenyl-beta-D-galactopyranoside (ONP-Gal) and 5-bromo-4-chloro-3-indolyl-beta-D-galactoside (X-gal). This A4 beta-galactosidase middle domain lacks the catalytic triad of typical GATase1 domains. The reactive Cys residue found in the sharp turn between a beta strand and an alpha helix termed the nucleophile elbow in typical GATase1 domains is not conserved in this group. |
| pfam16657 | Malt_amylase_C | 2.77e-07 | 1002 | 1067 | 4 | 75 | Maltogenic Amylase, C-terminal domain. This is the C-terminal domain of Maltogenic amylase, an enzyme that hydrolyzes starch material. Maltogenic amylases are central to carbohydrate metabolism. |
| pfam02449 | Glyco_hydro_42 | 1.07e-06 | 478 | 634 | 72 | 234 | Beta-galactosidase. This group of beta-galactosidase enzymes belong to the glycosyl hydrolase 42 family. The enzyme catalyzes the hydrolysis of terminal, non-reducing terminal beta-D-galactosidase residues. |
| COG1874 | GanA | 1.26e-06 | 478 | 854 | 94 | 487 | Beta-galactosidase GanA [Carbohydrate transport and metabolism]. |
| pfam00150 | Cellulase | 1.35e-06 | 72 | 290 | 29 | 252 | Cellulase (glycosyl hydrolase family 5). |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM43319.1 | 0.0 | 1 | 1072 | 1 | 1072 |
| AHF93474.1 | 1.91e-90 | 395 | 1033 | 79 | 729 |
| AGB50733.1 | 1.52e-23 | 70 | 328 | 49 | 327 |
| QHW30800.1 | 2.34e-22 | 401 | 858 | 893 | 1355 |
| QJD86938.1 | 5.23e-22 | 460 | 858 | 63 | 489 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 5XB7_A | 2.12e-10 | 414 | 865 | 26 | 502 | GH42alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis],5XB7_B GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis],5XB7_C GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis],5XB7_D GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis],5XB7_E GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis],5XB7_F GH42 alpha-L-arabinopyranosidase from Bifidobacterium animalis subsp. lactis Bl-04 [Bifidobacterium animalis subsp. lactis] |
| 4ZN2_A | 1.14e-09 | 70 | 295 | 36 | 261 | Glycosylhydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_B Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_C Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],4ZN2_D Glycosyl hydrolase from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1] |
| 5BX9_A | 1.14e-09 | 70 | 295 | 36 | 261 | Structureof PslG from Pseudomonas aeruginosa [Pseudomonas aeruginosa PAO1],5BXA_A Structure of PslG from Pseudomonas aeruginosa in complex with mannose [Pseudomonas aeruginosa PAO1] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q93GI5 | 6.14e-07 | 520 | 897 | 140 | 546 | Beta-galactosidase III OS=Bifidobacterium longum subsp. infantis OX=1682 GN=beta-galIII PE=1 SV=1 |
| A1A399 | 1.07e-06 | 416 | 855 | 37 | 497 | Beta-galactosidase BgaB OS=Bifidobacterium adolescentis (strain ATCC 15703 / DSM 20083 / NCTC 11814 / E194a) OX=367928 GN=bgaB PE=1 SV=2 |
SignalP and Lipop Annotations help
This protein is predicted as SP
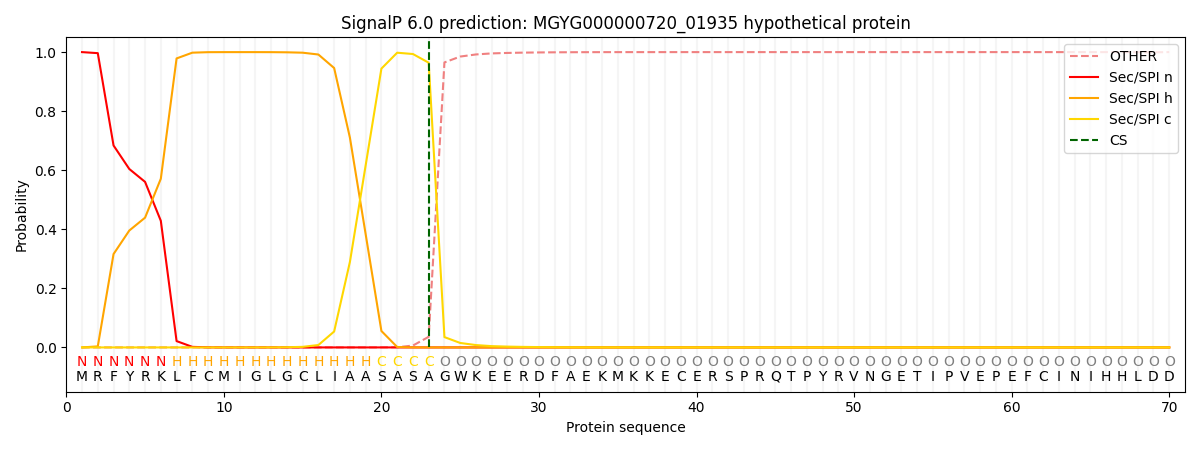
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001431 | 0.997525 | 0.000387 | 0.000230 | 0.000206 | 0.000210 |