You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_02241
You are here: Home > Sequence: MGYG000000720_02241
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_02241 | |||||||||||
| CAZy Family | GH2 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 953; End: 4477 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH2 | 512 | 914 | 4.2e-35 | 0.44813829787234044 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG3250 | LacZ | 2.49e-14 | 533 | 851 | 106 | 407 | Beta-galactosidase/beta-glucuronidase [Carbohydrate transport and metabolism]. |
| pfam02836 | Glyco_hydro_2_C | 2.14e-06 | 724 | 941 | 3 | 209 | Glycosyl hydrolases family 2, TIM barrel domain. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
| PRK10150 | PRK10150 | 7.37e-06 | 680 | 889 | 240 | 446 | beta-D-glucuronidase; Provisional |
| pfam00703 | Glyco_hydro_2 | 8.23e-06 | 610 | 720 | 1 | 106 | Glycosyl hydrolases family 2. This family contains beta-galactosidase, beta-mannosidase and beta-glucuronidase activities. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM44993.1 | 0.0 | 1 | 1174 | 54 | 1227 |
| SEH75953.1 | 9.96e-22 | 512 | 942 | 262 | 668 |
| QUH31336.1 | 8.21e-17 | 612 | 950 | 170 | 467 |
| AIQ32428.1 | 4.31e-16 | 533 | 950 | 102 | 518 |
| APU68349.1 | 7.10e-16 | 512 | 903 | 89 | 479 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3FN9_A | 4.05e-10 | 533 | 852 | 84 | 402 | Crystalstructure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_B Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_C Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343],3FN9_D Crystal structure of putative beta-galactosidase from bacteroides fragilis [Bacteroides fragilis NCTC 9343] |
| 4CU6_A | 2.36e-07 | 682 | 920 | 253 | 492 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU7_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4],4CU8_A Unravelling the multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA [Streptococcus pneumoniae TIGR4] |
| 4CUC_A | 2.36e-07 | 682 | 920 | 253 | 492 | Unravellingthe multiple functions of the architecturally intricate Streptococcus pneumoniae beta-galactosidase, BgaA. [Streptococcus pneumoniae TIGR4] |
| 6ED1_A | 1.22e-06 | 664 | 897 | 246 | 458 | ChainA, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_B Chain B, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_C Chain C, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei],6ED1_D Chain D, Glycosyl hydrolase family 2, sugar binding domain protein [Phocaeicola dorei] |
| 6NCW_A | 1.75e-06 | 533 | 886 | 73 | 404 | Crystalstructure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_B Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_C Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCW_D Crystal structure of a GH2 beta-galacturonidase from Eisenbergiella tayi bound to glycerol [Eisenbergiella tayi],6NCX_A Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_B Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_C Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi],6NCX_D Crystal structure of GH2 beta-galacturonidase from Eisenbergiella tayi bound to galacturonate [Eisenbergiella tayi] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| B2VHN8 | 7.99e-07 | 531 | 909 | 144 | 502 | Beta-galactosidase OS=Erwinia tasmaniensis (strain DSM 17950 / CFBP 7177 / CIP 109463 / NCPPB 4357 / Et1/99) OX=465817 GN=lacZ PE=3 SV=1 |
| Q48846 | 1.95e-06 | 533 | 892 | 153 | 492 | Beta-galactosidase large subunit OS=Latilactobacillus sakei OX=1599 GN=lacL PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
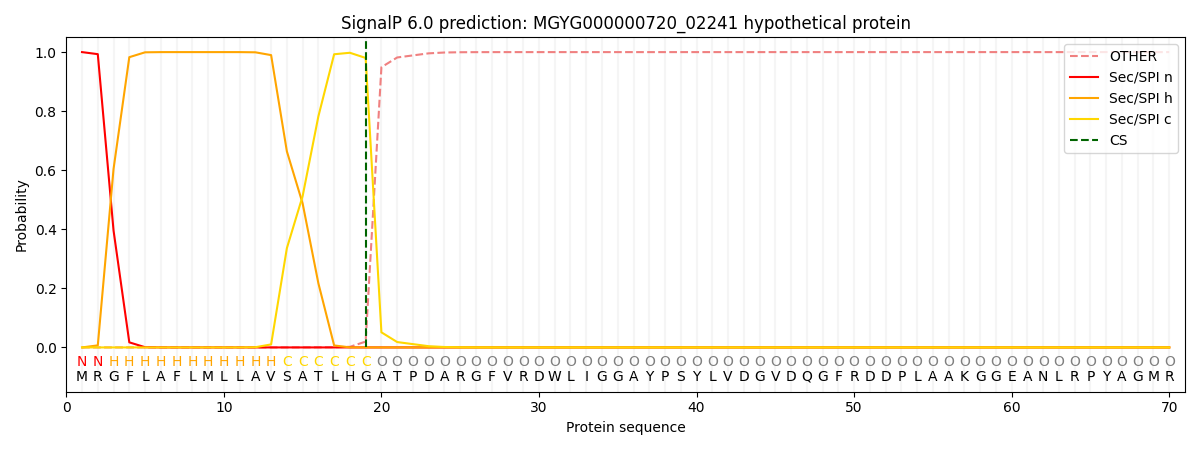
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000220 | 0.999151 | 0.000168 | 0.000160 | 0.000153 | 0.000143 |
