You are browsing environment: HUMAN GUT
MGYG000000720_02427
Basic Information
help
Species
Lineage
Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis;
CAZyme ID
MGYG000000720_02427
CAZy Family
GH51
CAZyme Description
hypothetical protein
CAZyme Property
Protein Length
CGC
Molecular Weight
Isoelectric Point
697
78970.13
7.0378
Genome Property
Genome Assembly ID
Genome Size
Genome Type
Country
Continent
MGYG000000720
3759035
MAG
Kazakhstan
Asia
Gene Location
Start: 13450;
End: 15543
Strand: -
No EC number prediction in MGYG000000720_02427.
Family
Start
End
Evalue
family coverage
GH51
304
689
4.4e-20
0.6079365079365079
Cdd ID
Domain
E-Value
qStart
qEnd
sStart
sEnd
Domain Description
smart00813
Alpha-L-AF_C
0.002
563
665
59
162
Alpha-L-arabinofuranosidase C-terminus. This entry represents the C terminus (approximately 200 residues) of bacterial and eukaryotic alpha-L-arabinofuranosidase. This catalyses the hydrolysis of non-reducing terminal alpha-L-arabinofuranosidic linkages in L-arabinose-containing polysaccharides.
more
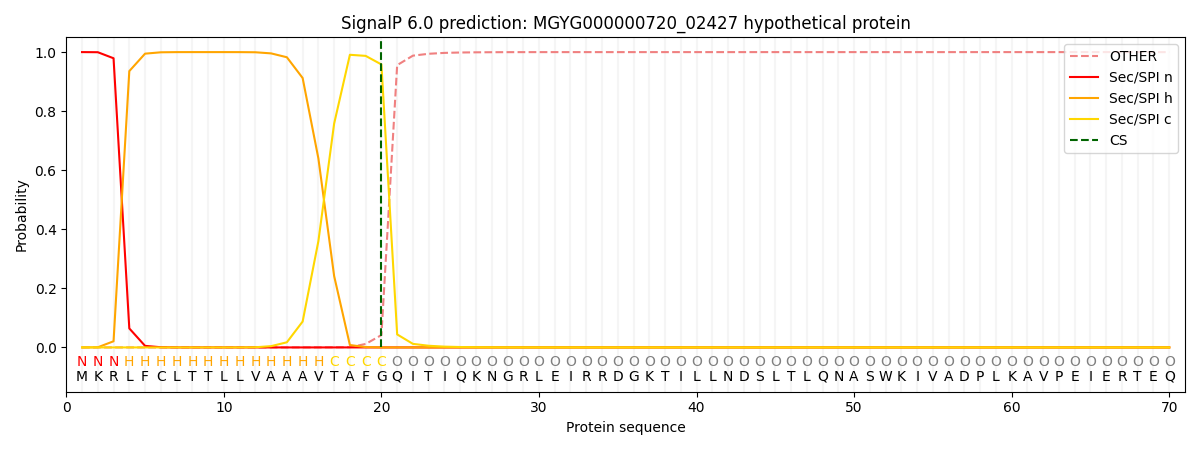
This protein is predicted as SP
Other
SP_Sec_SPI
LIPO_Sec_SPII
TAT_Tat_SPI
TATLIP_Sec_SPII
PILIN_Sec_SPIII
0.000395
0.998882
0.000232
0.000155
0.000156
0.000151