You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000720_03038
You are here: Home > Sequence: MGYG000000720_03038
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Verrucomicrobiota; Lentisphaeria; Victivallales; Victivallaceae; Victivallis; | |||||||||||
| CAZyme ID | MGYG000000720_03038 | |||||||||||
| CAZy Family | GH87 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 967; End: 2610 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH87 | 17 | 398 | 1.3e-19 | 0.5309882747068677 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 8.69e-16 | 18 | 275 | 79 | 362 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam12708 | Pectate_lyase_3 | 4.23e-12 | 23 | 189 | 3 | 141 | Pectate lyase superfamily protein. This family of proteins possesses a beta helical structure like Pectate lyase. This family is most closely related to glycosyl hydrolase family 28. |
| PLN02155 | PLN02155 | 3.03e-04 | 4 | 214 | 6 | 221 | polygalacturonase |
| PLN03003 | PLN03003 | 0.003 | 4 | 61 | 8 | 64 | Probable polygalacturonase At3g15720 |
| pfam13229 | Beta_helix | 0.003 | 112 | 225 | 8 | 107 | Right handed beta helix region. This region contains a parallel beta helix region that shares some similarity with Pectate lyases. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| AVM46992.1 | 7.34e-199 | 14 | 534 | 17 | 538 |
| AVM46991.1 | 2.87e-103 | 14 | 455 | 17 | 496 |
| BBY49293.1 | 2.83e-76 | 22 | 398 | 32 | 414 |
| AKK29103.1 | 1.43e-71 | 22 | 368 | 19 | 374 |
| BBZ59707.1 | 8.08e-69 | 1 | 368 | 7 | 393 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 2PYG_A | 1.92e-07 | 23 | 66 | 4 | 47 | Azotobactervinelandii Mannuronan C-5 epimerase AlgE4 A-module [Azotobacter vinelandii],2PYG_B Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module [Azotobacter vinelandii],2PYH_A Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide [Azotobacter vinelandii],2PYH_B Azotobacter vinelandii Mannuronan C-5 epimerase AlgE4 A-module complexed with mannuronan trisaccharide [Azotobacter vinelandii] |
| 5LW3_A | 4.61e-07 | 23 | 83 | 4 | 62 | Azotobactervinelandii Mannuronan C-5 epimerase AlgE6 A-module [Azotobacter vinelandii] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| Q44492 | 1.29e-08 | 23 | 307 | 4 | 265 | Mannuronan C5-epimerase AlgE5 OS=Azotobacter vinelandii OX=354 GN=algE5 PE=2 SV=1 |
| Q44494 | 7.33e-08 | 23 | 83 | 4 | 62 | Mannuronan C5-epimerase AlgE1 OS=Azotobacter vinelandii OX=354 GN=algE1 PE=1 SV=1 |
| Q44495 | 8.90e-08 | 23 | 83 | 4 | 62 | Mannuronan C5-epimerase AlgE2 OS=Azotobacter vinelandii OX=354 GN=algE2 PE=1 SV=1 |
| Q44496 | 2.29e-07 | 23 | 83 | 4 | 62 | Mannuronan C5-epimerase AlgE3 OS=Azotobacter vinelandii OX=354 GN=algE3 PE=3 SV=1 |
| Q44493 | 1.45e-06 | 23 | 66 | 4 | 47 | Mannuronan C5-epimerase AlgE4 OS=Azotobacter vinelandii OX=354 GN=algE4 PE=1 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
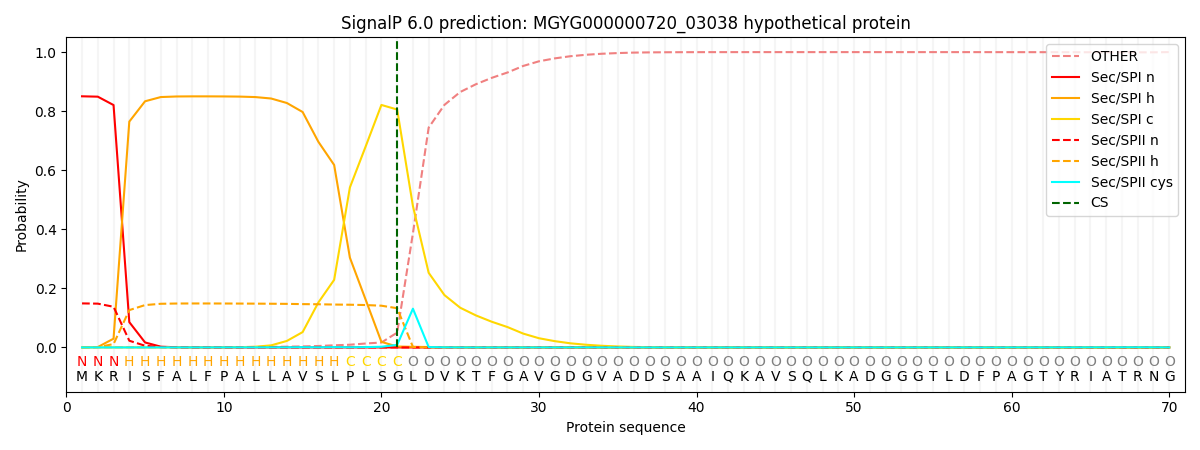
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.001715 | 0.845731 | 0.151729 | 0.000262 | 0.000263 | 0.000261 |
