You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000722_01181
You are here: Home > Sequence: MGYG000000722_01181
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | RC9 sp900545245 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Bacteroidota; Bacteroidia; Bacteroidales; UBA932; RC9; RC9 sp900545245 | |||||||||||
| CAZyme ID | MGYG000000722_01181 | |||||||||||
| CAZy Family | GH76 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 32951; End: 34429 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH76 | 123 | 461 | 2.6e-41 | 0.7821229050279329 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam03663 | Glyco_hydro_76 | 1.40e-15 | 180 | 387 | 86 | 255 | Glycosyl hydrolase family 76. Family of alpha-1,6-mannanases. |
| COG4833 | COG4833 | 3.61e-06 | 180 | 420 | 92 | 277 | Predicted alpha-1,6-mannanase, GH76 family [Carbohydrate transport and metabolism]. |
| cd01810 | Ubl2_ISG15 | 0.001 | 170 | 214 | 27 | 61 | ubiquitin-like (Ubl) domain 2 found in interferon-stimulated gene 15 (ISG15) and similar proteins. ISG15, also termed interferon-induced 15 kDa protein, or interferon-induced 17 kDa protein (IP17), or ubiquitin cross-reactive protein (UCRP), is an antiviral interferon-induced ubiquitin-like protein that upon viral infection it modifies cellular and viral proteins by mechanisms similar to ubiquitination. Although ISG15 has properties similar to those of other ubiquitin-like (Ubl) molecules, it is a unique member of the Ubl superfamily, whose expression and conjugation to target proteins are tightly regulated by specific signaling pathways, indicating it may have specialized functions in the immune system. ISG15 contains two tandem Ubl domains with a beta-grasp Ubl fold. This family corresponds to the second Ubl domain. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QIK59424.1 | 3.67e-185 | 8 | 490 | 4 | 478 |
| QIK53975.1 | 1.39e-182 | 8 | 490 | 4 | 478 |
| AGA76828.1 | 1.77e-181 | 9 | 490 | 8 | 481 |
| QDH77939.1 | 1.44e-180 | 9 | 490 | 11 | 481 |
| AWW30266.1 | 4.72e-179 | 9 | 490 | 11 | 481 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MU9_A | 4.51e-17 | 166 | 465 | 105 | 347 | Crystalstructure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482],4MU9_B Crystal structure of a putative glycosylhydrolase (BT_3782) from Bacteroides thetaiotaomicron VPI-5482 at 1.89 A resolution [Bacteroides thetaiotaomicron VPI-5482] |
| 4C1S_A | 5.19e-16 | 54 | 444 | 6 | 329 | Glycosidehydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482],4C1S_B Glycoside hydrolase family 76 (mannosidase) Bt3792 from Bacteroides thetaiotaomicron VPI-5482 [Bacteroides thetaiotaomicron VPI-5482] |
| 6SHD_A | 5.91e-11 | 177 | 415 | 130 | 318 | Structureof the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_B Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6],6SHD_C Structure of the GH76A alpha-1,6-mannanase from Salegentibacter sp. HEL1_6 [Salegentibacter sp. Hel_I_6] |
| 6Y8F_A | 1.03e-09 | 177 | 415 | 131 | 319 | ChainA, Alpha-1,6-endo-mannanase GH76A mutant [Salegentibacter sp. Hel_I_6] |
| 6SHM_A | 5.65e-09 | 170 | 415 | 116 | 319 | Aninactive (D136A and D137A) variant of alpha-1,6-mannanase, GH76A of Salegentibacter sp. HEL1_6 in complex with alpha-1,6-mannotetrose [Salegentibacter sp. Hel_I_6] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as LIPO
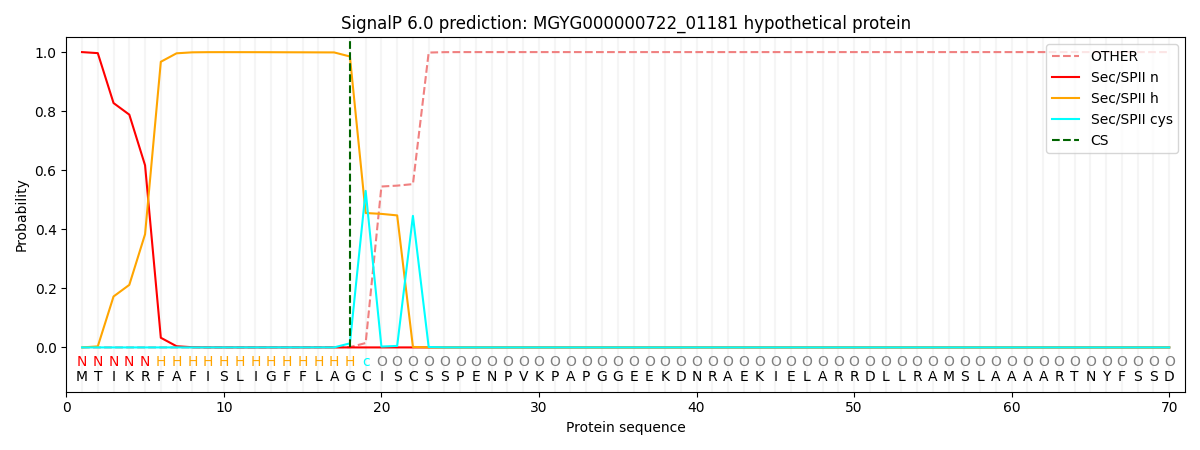
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.000001 | 0.000033 | 1.000010 | 0.000000 | 0.000000 | 0.000000 |
