You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000729_01571
You are here: Home > Sequence: MGYG000000729_01571
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
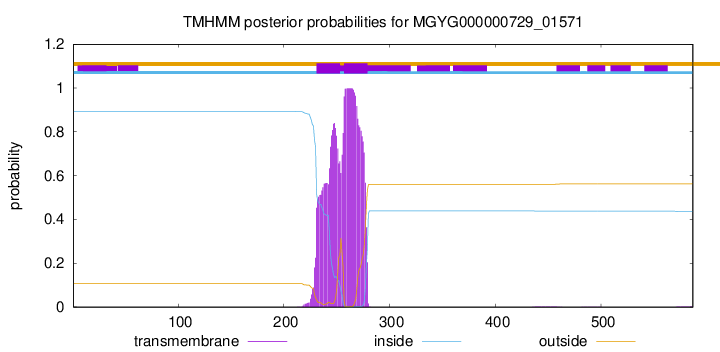
TMHMM annotations
Basic Information help
| Species | F23-B02 sp900545805 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia; Oscillospirales; Oscillospiraceae; F23-B02; F23-B02 sp900545805 | |||||||||||
| CAZyme ID | MGYG000000729_01571 | |||||||||||
| CAZy Family | GH23 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 47538; End: 49301 Strand: - | |||||||||||
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| pfam05257 | CHAP | 6.37e-15 | 472 | 562 | 1 | 83 | CHAP domain. This domain corresponds to an amidase function. Many of these proteins are involved in cell wall metabolism of bacteria. This domain is found at the N-terminus of Escherichia coli gss, where it functions as a glutathionylspermidine amidase EC:3.5.1.78. This domain is found to be the catalytic domain of PlyCA. CHAP is the amidase domain of bifunctional Escherichia coli glutathionylspermidine synthetase/amidase, and it catalyzes the hydrolysis of Gsp (glutathionylspermidine) into glutathione and spermidine. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| VCV20591.1 | 3.15e-142 | 1 | 587 | 1 | 591 |
| CBL08623.1 | 4.44e-142 | 1 | 587 | 1 | 591 |
| CCO05014.1 | 8.87e-142 | 1 | 587 | 1 | 591 |
| QNM10177.1 | 3.98e-140 | 1 | 587 | 1 | 591 |
| CBL14462.1 | 1.12e-139 | 1 | 587 | 1 | 591 |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as OTHER
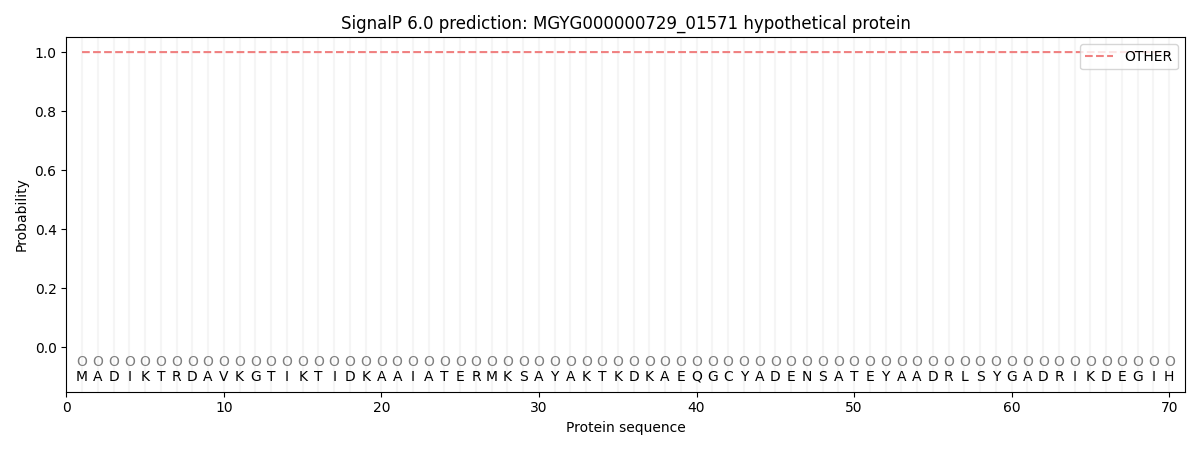
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 1.000027 | 0.000001 | 0.000000 | 0.000000 | 0.000000 | 0.000000 |