You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000733_00076
You are here: Home > Sequence: MGYG000000733_00076
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
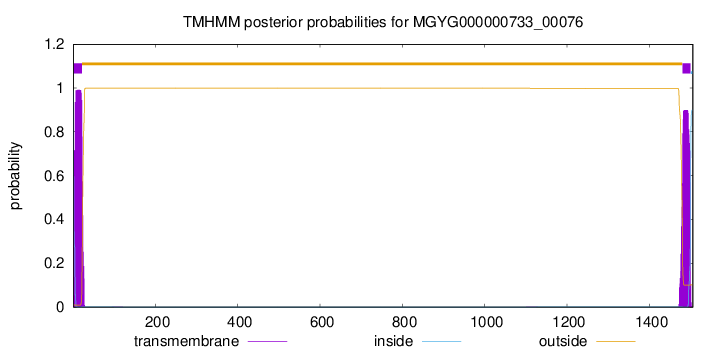
TMHMM annotations
Basic Information help
| Species | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; Borkfalkia; | |||||||||||
| CAZyme ID | MGYG000000733_00076 | |||||||||||
| CAZy Family | CE1 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 3914; End: 8434 Strand: + | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| CE1 | 972 | 1138 | 1.1e-26 | 0.5947136563876652 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG4099 | COG4099 | 6.31e-29 | 928 | 1190 | 134 | 383 | Predicted peptidase [General function prediction only]. |
| COG3509 | LpqC | 1.57e-11 | 933 | 1118 | 17 | 177 | Poly(3-hydroxybutyrate) depolymerase [Secondary metabolites biosynthesis, transport and catabolism]. |
| NF033189 | internalin_A | 3.64e-11 | 1223 | 1469 | 460 | 704 | class 1 internalin InlA. Internalins, as found in the intracellular human pathogen Listeria monocytogenes, are paralogous surface or secreted proteins with an N-terminal signal peptide, leucine-rich repeats, and usually a C-terminal LPXTG processing and cell surface anchoring site. See PMID:17764999 for a general discussion of internalins. Members of this family are internalin A (InlA), a class 1 (LPXTG-type) internalin. |
| pfam09479 | Flg_new | 3.94e-11 | 1418 | 1465 | 13 | 64 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| pfam09479 | Flg_new | 8.05e-09 | 1197 | 1263 | 2 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| BCI61582.1 | 1.39e-35 | 748 | 1347 | 615 | 1174 |
| ABS60377.1 | 7.67e-26 | 970 | 1194 | 20 | 245 |
| VTR91196.1 | 4.30e-20 | 963 | 1194 | 34 | 239 |
| QDU56037.1 | 8.67e-20 | 893 | 1194 | 712 | 1007 |
| ACR12533.1 | 1.93e-19 | 948 | 1201 | 40 | 288 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 3DOH_A | 4.67e-28 | 964 | 1194 | 150 | 380 | CrystalStructure of a Thermostable Esterase [Thermotoga maritima],3DOH_B Crystal Structure of a Thermostable Esterase [Thermotoga maritima],3DOI_A Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima],3DOI_B Crystal Structure of a Thermostable Esterase complex with paraoxon [Thermotoga maritima] |
| 3WYD_A | 1.12e-17 | 972 | 1194 | 21 | 217 | C-terminalesterase domain of LC-Est1 [uncultured organism],3WYD_B C-terminal esterase domain of LC-Est1 [uncultured organism] |
Swiss-Prot Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| P0DJ98 | 7.10e-20 | 1258 | 1468 | 1 | 221 | Putative membrane protein Bcell_0381 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0381 PE=4 SV=1 |
| P0DJ97 | 1.34e-19 | 1228 | 1475 | 465 | 744 | Putative Gly-rich membrane protein Bcell_0380 OS=Evansella cellulosilytica (strain ATCC 21833 / DSM 2522 / FERM P-1141 / JCM 9156 / N-4) OX=649639 GN=Bcell_0380 PE=4 SV=1 |
| P0DJM0 | 1.45e-09 | 1273 | 1469 | 502 | 705 | Internalin A OS=Listeria monocytogenes serovar 1/2a (strain ATCC BAA-679 / EGD-e) OX=169963 GN=inlA PE=1 SV=1 |
| Q723K6 | 1.45e-09 | 1273 | 1469 | 502 | 705 | Internalin A OS=Listeria monocytogenes serotype 4b (strain F2365) OX=265669 GN=inlA PE=3 SV=1 |
SignalP and Lipop Annotations help
This protein is predicted as SP
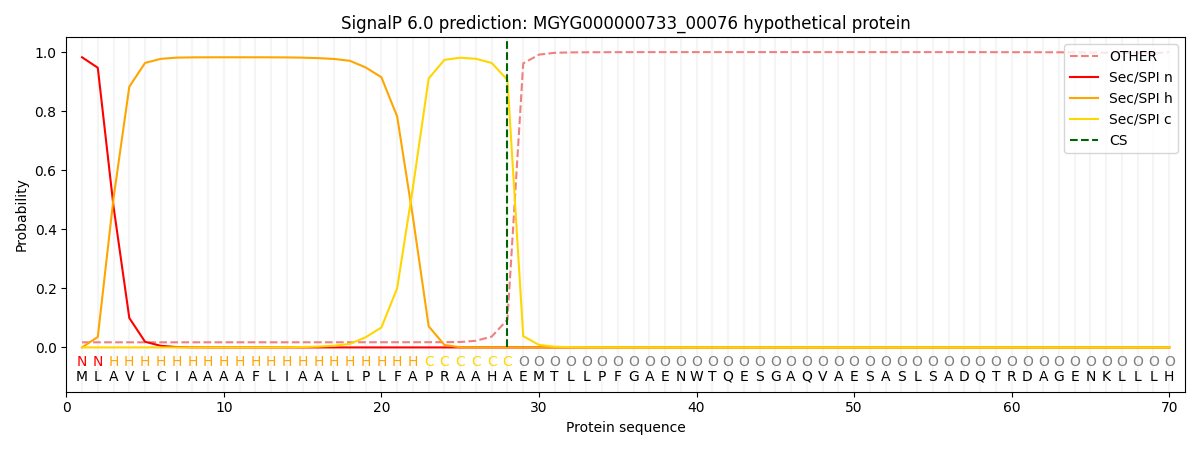
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.034743 | 0.962028 | 0.001019 | 0.001180 | 0.000541 | 0.000464 |