You are browsing environment: HUMAN GUT
CAZyme Information: MGYG000000734_00730
You are here: Home > Sequence: MGYG000000734_00730
Basic Information |
Genomic context |
Full Sequence |
Enzyme annotations |
CAZy signature domains |
CDD domains |
CAZyme hits |
PDB hits |
Swiss-Prot hits |
SignalP and Lipop annotations |
TMHMM annotations
Basic Information help
| Species | UBA10281 sp900548285 | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Lineage | Bacteria; Firmicutes_A; Clostridia_A; Christensenellales; Borkfalkiaceae; UBA10281; UBA10281 sp900548285 | |||||||||||
| CAZyme ID | MGYG000000734_00730 | |||||||||||
| CAZy Family | GH28 | |||||||||||
| CAZyme Description | hypothetical protein | |||||||||||
| CAZyme Property |
|
|||||||||||
| Genome Property |
|
|||||||||||
| Gene Location | Start: 61189; End: 71055 Strand: - | |||||||||||
CAZyme Signature Domains help
| Family | Start | End | Evalue | family coverage |
|---|---|---|---|---|
| GH28 | 2525 | 2775 | 7.6e-21 | 0.5876923076923077 |
CDD Domains download full data without filtering help
| Cdd ID | Domain | E-Value | qStart | qEnd | sStart | sEnd | Domain Description |
|---|---|---|---|---|---|---|---|
| COG5434 | Pgu1 | 3.00e-21 | 2467 | 2773 | 76 | 369 | Polygalacturonase [Carbohydrate transport and metabolism]. |
| pfam09479 | Flg_new | 1.37e-08 | 3164 | 3229 | 1 | 65 | Listeria-Bacteroides repeat domain (List_Bact_rpt). This model describes a conserved core region of about 43 residues, which occurs in at least two families of tandem repeats. These include 78-residue repeats which occur from 2 to 15 times in some proteins of Bacteroides forsythus ATCC 43037, and 70-residue repeats found in families of internalins of Listeria species. Single copies are found in proteins of Fibrobacter succinogenes, Geobacter sulfurreducens, and a few other bacteria. |
| pfam00295 | Glyco_hydro_28 | 7.81e-05 | 2581 | 2777 | 48 | 214 | Glycosyl hydrolases family 28. Glycosyl hydrolase family 28 includes polygalacturonase EC:3.2.1.15 as well as rhamnogalacturonase A(RGase A), EC:3.2.1.-. These enzymes are important in cell wall metabolism. |
| pfam02368 | Big_2 | 7.44e-04 | 1791 | 1859 | 8 | 76 | Bacterial Ig-like domain (group 2). This family consists of bacterial domains with an Ig-like fold. Members of this family are found in bacterial and phage surface proteins such as intimins. |
| pfam18998 | Flg_new_2 | 0.002 | 3185 | 3230 | 22 | 70 | Divergent InlB B-repeat domain. This family of domains are found in bacterial cell surface proteins. They are often found in tandem array. This domain is closely related to pfam09479. |
CAZyme Hits help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End |
|---|---|---|---|---|---|
| QGA23464.1 | 5.87e-159 | 2210 | 2951 | 71 | 794 |
| QCD41820.1 | 3.03e-104 | 2255 | 2913 | 246 | 884 |
| QGQ94767.1 | 2.63e-38 | 2470 | 2970 | 34 | 533 |
| ADB48561.1 | 2.16e-36 | 2470 | 3002 | 25 | 531 |
| ADB48560.1 | 1.79e-34 | 2470 | 2967 | 25 | 516 |
PDB Hits download full data without filtering help
| Hit ID | E-Value | Query Start | Query End | Hit Start | Hit End | Description |
|---|---|---|---|---|---|---|
| 4MXN_A | 7.20e-11 | 2480 | 2648 | 27 | 171 | Crystalstructure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_B Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_C Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184],4MXN_D Crystal structure of a putative glycosyl hydrolase (PARMER_00599) from Parabacteroides merdae ATCC 43184 at 1.95 A resolution [Parabacteroides merdae ATCC 43184] |
Swiss-Prot Hits help
SignalP and Lipop Annotations help
This protein is predicted as SP
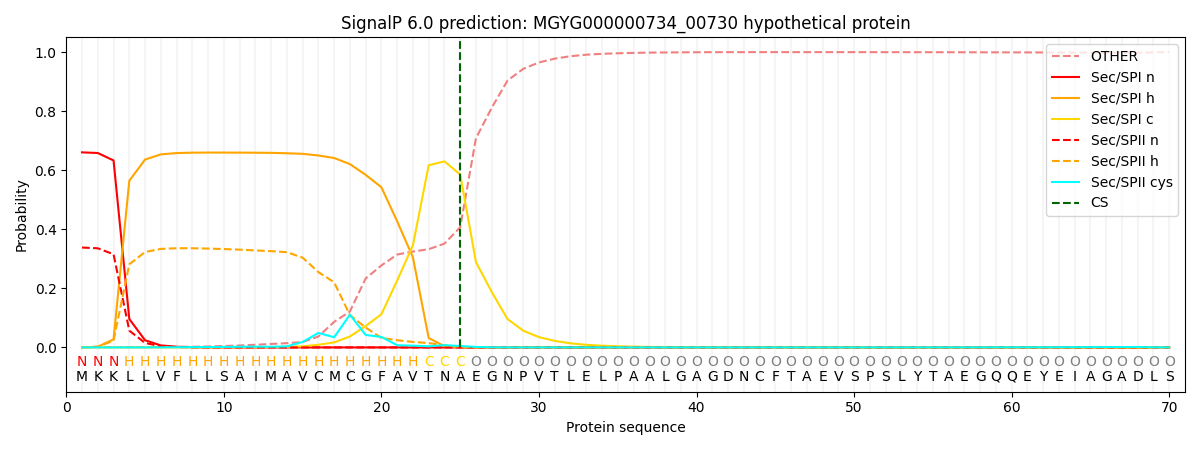
| Other | SP_Sec_SPI | LIPO_Sec_SPII | TAT_Tat_SPI | TATLIP_Sec_SPII | PILIN_Sec_SPIII |
|---|---|---|---|---|---|
| 0.002239 | 0.650793 | 0.345998 | 0.000394 | 0.000289 | 0.000257 |
TMHMM Annotations download full data without filtering help
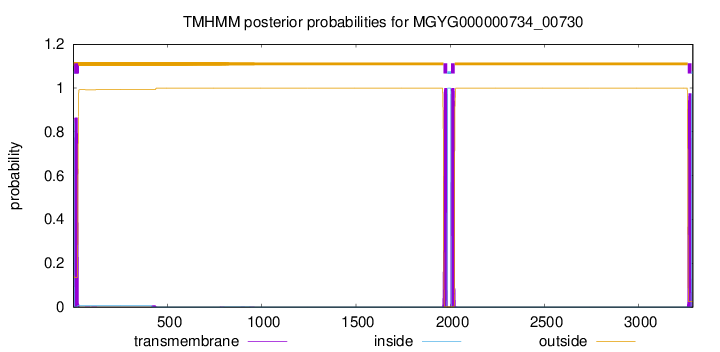
| start | end |
|---|---|
| 5 | 24 |
| 1965 | 1984 |
| 2004 | 2023 |
| 3261 | 3280 |
